Viagra gibt es mittlerweile nicht nur als Original, sondern auch in Form von Generika. Diese enthalten denselben Wirkstoff Sildenafil. Patienten suchen deshalb nach viagra generika schweiz, um ein günstigeres Präparat zu finden. Unterschiede bestehen oft nur in Verpackung und Preis.
Prodeinsteinwebsite.vipprod.dmz.yu.edu

Evolutionarily conserved gene family important
for fat storage
Bert Kadereit*, Pradeep Kumar*, Wen-Jun Wang*, Diego Miranda*, Erik L. Snapp†, Nadia Severina‡, Ingrid Torregroza‡,
Todd Evans‡, and David L. Silver*§
*Department of Biochemistry and the Marion Bessin Liver Research Center and Departments of ‡Developmental and Molecular Biology and †Anatomyand Structural Biology, Albert Einstein College of Medicine, Bronx, NY 10461
Edited by Kaveh Ashrafi, University of California, San Francisco, CA, and accepted by the Editorial Board November 11, 2007 (received for reviewSeptember 11, 2007)
The ability to store fat in the form of cytoplasmic triglyceride droplets
able to diacylglycerol acyltransferase enzymes), and are regu-
is conserved from Saccharomyces cerevisiae to humans. Although
lated by the peroxisome proliferator-activated receptor nuclear
much is known regarding the composition and catabolism of lipid
hormone receptors, PPAR␣ and PPAR␥, which play crucial
droplets, the molecular components necessary for the biogenesis of
roles in lipid metabolism. Here we describe the identification and
lipid droplets have remained obscure. Here we report the character-
characterization of a family of proteins that fit these character-
ization of a conserved gene family important for lipid droplet forma-
istics and are important for the accumulation of lipid droplets.
tion named fat-inducing transcript (FIT). FIT1 and FIT2 are endoplasmic
reticulum resident membrane proteins that induce lipid droplet ac-
cumulation in cell culture and when expressed in mouse liver. shRNA
Identification of the FIT Family. We sought to identify proteins
silencing of FIT2 in 3T3-LI adipocytes prevents accumulation of lipid
involved in intracellular fatty acid transport and metabolism either
droplets, and depletion of FIT2 in zebrafish blocks diet-induced
through storage as TG in lipid droplets or catabolism by -oxidation
accumulation of lipid droplets in the intestine and liver, highlighting
in mitochondria. Fenofibrate and other fibrate drugs are specific
an important role for FIT2 in lipid droplet formation in vivo. Together
agonists for the peroxisome proliferator-activated receptor-␣
these studies identify and characterize a conserved gene family that
(PPAR␣) nuclear hormone receptor, and activation of PPAR␣
is important in the fundamental process of storing fat.
leads to enhanced gene expression for much of the biochemicalrepertoire of -oxidation (16). Genotype matched WT and
adipocytes 兩 diabetes 兩 FIT 兩 obesity 兩 triglyceride
PPAR␣-deficient mice were fed a diet containing fenofibrate for 7days. RNA was purified from livers to generate cDNA probes used
The ability to store energy in the form of triglyceride (TG) is to query a gene array. Because many of the genes activated by
conserved from Saccharomyces cerevisiae to humans. TGs
PPAR␣ have been identified, we focused exclusively on genes that
are stored in the cytoplasm surrounded by a monolayer of
were listed as ESTs or having unknown function. Our attention was
phospholipid in distinct structures or organelles given numerous
directed toward two unknown transcripts: fat-inducing transcript 1
names, such as lipid particles, oil bodies, adiposomes, eicosa-
(FIT1) and FIT2. The fenofibrate-induced expression of both
somes, and, more commonly, lipid droplets (1). Under normal
mouse FIT1 and FIT2 was confirmed to be PPAR␣-dependent in
physiological conditions, lipid droplets are involved in maintain-
the liver and not induced by fenofibrate in the heart
ing energy balance at the cellular and whole-organism levels. Yet
FIT1 and FIT2 genes encode putative 292-
under conditions of extreme lipid droplet acquisition, as in
and 262 amino acid proteins, respectively, which are 50% similar to
obesity, the risk for acquiring common debilitating diseases such
each other (Fig. 1A). FIT1 and FIT2 contain multiple potential
as type 2 diabetes and cardiovascular diseases is increased (2).
transmembrane domains that are highly expressed in
Despite their central role in energy homeostasis, only recently
heart and skeletal muscle according to gene array studies (deter-
have the composition and functions of many of the components
mined by Novartis Gene Atlas). However, FIT1 and FIT2 do not
of lipid droplets from S. cerevisiae, Drosophila, and mammalian
have homology to known proteins or protein domains found in any
cells been revealed. In general, lipid droplets are composed of a
species, indicating that FIT genes comprise a unique gene family.
core of neutral lipids, primarily TGs, surrounded by a monolayer
These characteristics indicated that these proteins are potentially
of phospholipids and lipid droplet-associated proteins (3–7). In
involved in lipid metabolism in oxidative tissues.
mammalian cells, the catabolism of lipid droplets is a highly
Performing a BLAST search with the full-length mouse FIT1
regulated process involving hormonal signals, droplet-associated
amino acid sequence against the expressed database, we identified
proteins, and lipases (8–10). Although much has been learned
FIT1 and FIT2 orthologous genes in mammals, whereas only a
about the components and catabolism of lipid droplets, the
single FIT gene can be identified in amphibians, birds, insects, and
molecular mechanism of lipid droplet biogenesis has remained
worms, which exhibit higher homology to FIT2 (Fig. 1B). In
unknown. The prevailing view is that lipid droplets are formedat the endoplasmic reticulum (ER) because the ER is the site of
Author contributions: T.E. and D.L.S. designed research; B.K., P.K., W.-J.W., D.M., E.L.S., N.S.,
TG biosynthesis, and lipid droplets are often observed in close
I.T., and D.L.S. performed research; E.L.S. contributed new reagents/analytic tools; B.K. and
association with the cytoplasmic face of the ER (11–13). A
D.L.S. analyzed data; and D.L.S. wrote the paper.
widely accepted model of lipid droplet biogenesis involves the
The authors declare no conflict of interest.
formation of a core or lens of newly synthesized TG between the
This article is a PNAS Direct Submission. K.A. is a guest editor invited by the Editorial Board.
leaflets of the ER membrane that buds off with the cytoplasmic
Freely available online through the PNAS open access option.
leaflet of the ER surrounding the neutral lipid core and acquires
Data deposition: The data reported in this paper have been deposited in the Gene
exchangeable cytosolic lipid droplet-associated proteins (14).
However, this view was recently challenged by observations
GSM136099 and GSM136100).
suggesting that lipid droplets form on the cytosolic leaflet of the
§To whom correspondence should be addressed. E-mail: [email protected].
ER membrane (15). In any case, we surmised that proteins that
This article contains supporting information online at
mediate lipid droplet biogenesis are localized to the ER, are not
directly involved in the biosynthesis of TG (an activity attribut-
2007 by The National Academy of Sciences of the USA
94 –99 兩 PNAS 兩 January 8, 2008 兩 vol. 105 兩 no. 1
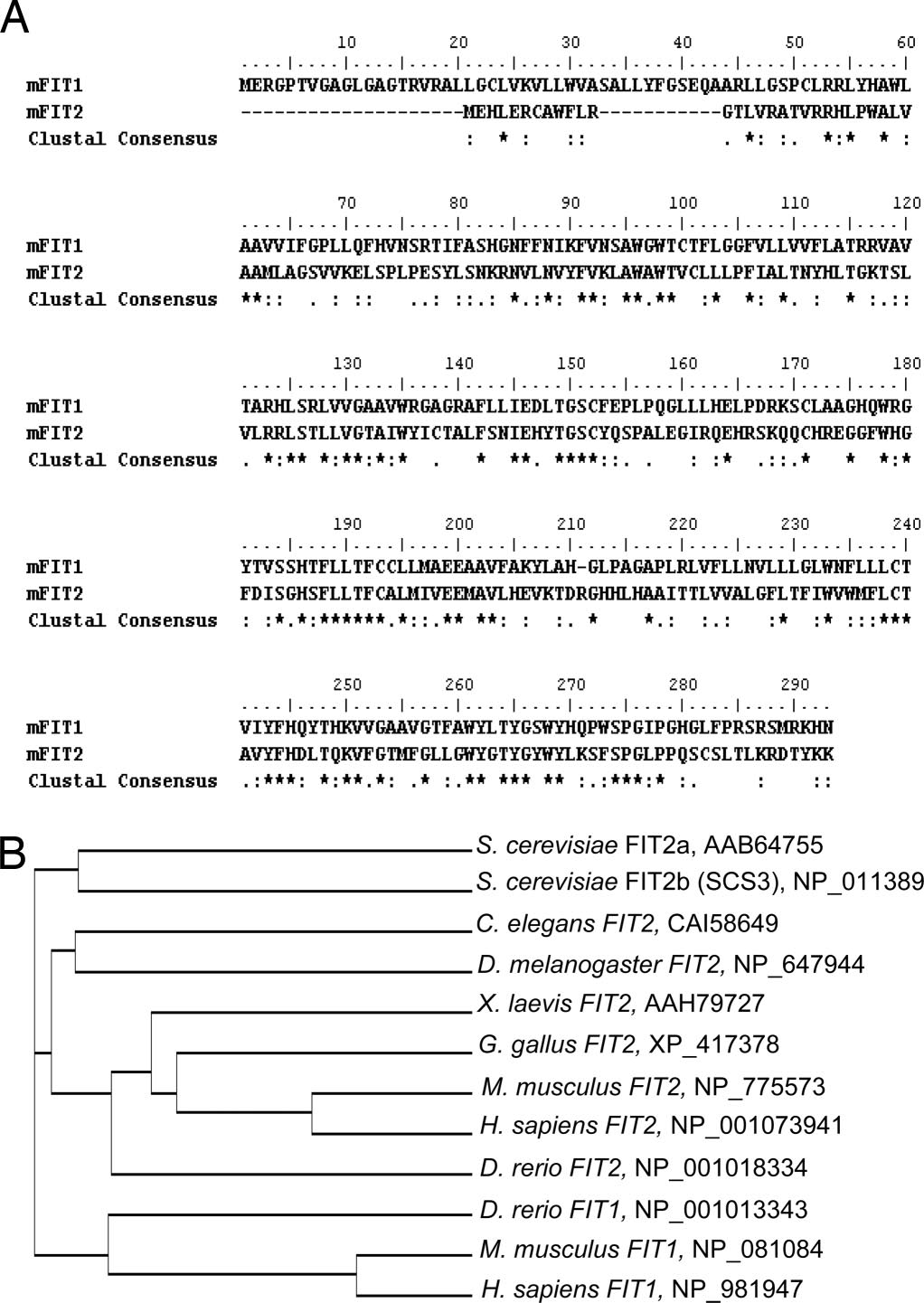

FITs Are ER Resident Membrane Proteins. Subcellular fractionation of
mouse heart membranes and confocal immunolocalization studies
were used to determine the subcellular localization of FITs. Frac-
tionation of mouse heart membranes by sucrose density ultracen-
trifugation indicated that both FIT1 and FIT2 colocalized with the
ER membrane resident protein Sec61- (Fig. 2D) and slightly
lighter membranes (fraction 9). The bands found in fractions 1 and
2 represent nonspecific antigens of the incorrect molecular weight
and were observed by using these polyclonal antibodies to FIT1 and
FIT2 in total cell lysates from mouse tissues and in HEK293 cells
(Fig. 2B). To exclude the possibility that FIT proteins also localize
to lipid droplets, we examined the localization of FIT2 in murine
brown adipose tissue, a major tissue in which FIT2 is expressed
(FIT1 is not expressed in adipose tissue or adipocytes in culture)
(Fig. 2 A). Separation of lipid droplets from membranes of brown
adipose tissue indicated that FIT2 does not reside in the lipid
droplet fraction enriched in TG and the lipid droplet-associated
PAT family protein, perilipin (Fig. 2E). Expression of carboxyl-
terminal V5 epitope-tagged FIT1 or FIT2 in HEK293 cells resulted
in a clear reticular staining pattern that colocalized with a synthetic
ER marker protein, red fluorescent protein (RFP) having the ER
retention signal, KDEL, but not at all with the Golgi-specific
marker protein, galactosyl-transferase-RFP (Fig. 2F). Also noted in
some cells expressing FITs was the presence of single, large,
oval-shaped fluorescent ER domains (colocalizing with ER-RFP)
likely representing proliferated smooth ER that commonly occurs
as a result of overexpression of ER membrane proteins (18, 19).
Together these findings provide further support for exclusive ER
localization of FIT proteins. The V5 epitope tag on FIT1 and FIT2
did not abolish its activity (see In agreement with the ER
localization of mouse FIT1 and FIT2, one of the S. cerevisiae FIT2
orthologs, FIT2b (FIT2a has not yet been localized), has been
localized exclusively to the ER in a high-throughput attempt to
localize all expressed ORFs in S. cerevisiae (20). Together the data
indicate that FIT1 and FIT2 are ER resident membrane proteins,
the established site for TG biosynthesis and the proposed site for
FIT sequence analysis. (A) Amino acid sequence alignment of murine
lipid droplet biogenesis.
FIT1 and FIT2 (35% identical, 50% similar). (B) Sequence alignments of FITorthologs in multiple species. Cladogram generated with ClustalW showing
FIT-Mediated Lipid Droplet Accumulation.
the amino acid sequence homologies among FIT proteins. Accession numbers
To determine whether FIT
for each FIT ortholog are indicated next to the cladogram.
proteins play a role in lipid metabolism, we overexpressed FITproteins in HEK293 cells and examined the presence of lipiddroplets by using the fluorescent lipid droplet stain (neutral lipid
contrast, the zebrafish genome contains orthologs of both mam-
stain) BODIPY493/503. Overexpression of FIT1 or FIT2, at similar
malian FIT1 and FIT2. S. cerevisiae has two FIT2 orthologs
levels resulted in the accumulation of lipid droplets (Fig.
(designated here as FIT2a and FIT2b/SCS3), which is consistent
3A). As a positive control, cells expressing DGAT1, one of two
with evidence that the ancestral S. cerevisiae underwent a genome-
acyltransferases important in the committed step in TG biosynthe-
wide duplication (17).
sis (11), showed multiple lipid droplets per cell (Fig. 3A). Compared
Northern blot analysis of mouse tissues indicated that FIT1 is
with mock-transfected cells, overexpression of DGAT1 or DGAT2,
highly expressed in heart and skeletal muscle, as well as at lower
a second acyltransferase important in the committed step in TG
levels in the liver, kidney, and testes (Fig. 2A). Western blot
biosynthesis (11), resulted in a 7- to 10-fold increase in levels of
analysis of mouse tissues indicated that FIT1 protein was de-
cellular TG, whereas TG levels increased only a modest 2-fold in
tected primarily in skeletal muscle, with lower levels in the heart
FIT1- and FIT2-expressing cells (Fig. 3B). Overexpression of FIT1
(Fig. 2B). Mouse FIT2 mRNA was ubiquitously detected as two
or FIT2 did not change the levels of phospholipids
to three transcripts, with highest levels in white and brown
Cholesteryl esters (CEs) in FIT1-, FIT2-, or DGAT2-overexpress-ing cells significantly increased, compared with control cells
adipose tissue, the heart, and skeletal muscle (Fig. 2 A). Western
Because CEs also increased in DGAT2-expressing cells and
blot analysis indicated that FIT2 protein was ubiquitously de-
DGAT2 does not use cholesterol as a substrate, being highly specific
tected, with highest levels in white and brown adipose tissue (Fig.
for diacyglycerol (12), we concluded that moderate increases in CEs
2B). The levels of mFIT1 and mFIT2 mRNA do not correlate
in HEK293 cells is an indirect consequence of lipid droplet
well with protein levels in the heart, indicating potential post-
transcriptional regulation in this organ. An examination of
We next tested whether FIT1 and FIT2 enhance TG biosynthesis
human tissues showed that FIT1 was primarily expressed in heart
by quantifying the rate of TG biosynthesis by using radiolabeled
and skeletal muscle, whereas FIT2 was expressed in all tissues
glycerol as a precursor for TG. Overexpression of positive controls
represented on the Northern blot (Fig. 2C). Together these
DGAT1 and DGAT2 led to a significant increase in the rate of TG
analyses indicated that both mouse and human FIT1 have an
biosynthesis (Fig. 3C) above mock-transfected cells as previously
expression pattern more restricted to oxidative tissues, whereas
shown (11, 12). In contrast, cells expressing FIT1 or FIT2 resulted
FIT2 has a broader expression pattern, with highest levels in
in similar rates of TG biosynthesis, compared with mock-
mouse adipose tissue (human adipose tissue was not examined).
transfected cells (Fig. 3C). In addition, the mRNAs of genes
Kadereit et al.
PNAS 兩 January 8, 2008 兩 vol. 105 兩 no. 1 兩 95


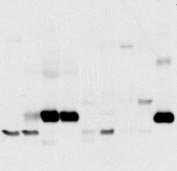
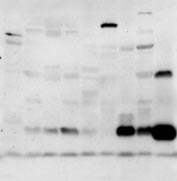
Analysis of FIT expression and localization. (A) Fifteen micrograms of total RNA from the mouse tissues shown was subjected to Northern blot analysis
for murine FIT1 and FIT2 (mfit1 and mfit2). Sk muscle, skeletal muscle; wat, white adipose tissue; bat, brown adipose tissue. The ethidium bromide-stained gelindicates loading. (B) Eighty micrograms of total cell lysates from the selected mouse tissues shown was subjected to Western blot analysis. Calnexin served asa loading control. Other cross-reacting bands not indicated by arrows are nonspecific. Lysates from HEK293 cells expressing FIT1 or FIT2 served as positive controls.
(C) A human RNA blot was analyzed for both fit1 and fit2 (hfit1 and hfit2) expression. (D) Total postnuclear membranes from mouse hearts were separated bycontinuous sucrose gradients (fractions shown from lowest to highest density). Fractions were subjected to Western blot analysis by using antibody markers forthe plasma membrane (Na K-ATPase), Golgi apparatus (FTCD/Gogli 58-kDa protein), ER (Sec61-), and FIT1 and FIT2. (E) Lipid droplets and membranes frommouse brown adipose tissue were fractionated on a continuous sucrose gradient, and fractions were subjected to Western blot analysis for FIT2 and perilipin(Plpn) and determination of TG in each fraction by TLC analysis. (F) Mouse FIT1-V5 and FIT2-V5 colocalized with the ER marker protein RFP-KDEL (ER-RFP) butnot with the Golgi-specific marker GalTase-RFP (Golgi-RFP) in HEK293 cells. (Scale bar: 10 m.)
important in fatty acid and TG biosynthesis were not changed in
in the liver (Fig. 2B). The histological examination of livers from
cells expressing FIT1 or FIT2 Taken together, the data
FIT2 adenovirus-injected mice indicated increased lipid droplets,
indicate that FIT1 and FIT2 do not enhance TG biosynthesis, but
compared with control livers (arrows indicate lipid droplets) (Fig.
rather increase the partitioning of cellular TG into lipid droplets. To
4A). In agreement with the appearance of increased lipid droplets,
provide evidence that FIT1 and FIT2 increase the partitioning of
FIT2-overexpressing livers (Fig. 4C) had a significant increase in
TG into lipid droplets, HEK293 cells were transfected with FIT1 or
TG, but normal levels of cholesterol, compared with controls
FIT2 and treated for 3 h with radiolabeled glycerol to label newly
(Fig. 4B).
synthesized TG; then lipid droplets were isolated by ultracentrifu-gation and radioactive TG-quantified. Expression of FIT1 and FIT2
shRNA Knockdown of FIT2 in Adipocytes. The requirement of FIT
significantly increased (between 4- and 6-fold) the amount of
proteins for lipid droplet biogenesis was investigated by using
labeled, newly synthesized TG in lipid droplets, compared with
shRNA technology. We surmised that if FIT proteins were essential
control cells (Fig. 3D), supporting the conclusion that FIT1 and
for lipid droplet formation, then FITs should be expressed during
FIT2 do not affect TG biosynthesis (Fig. 3C), but rather partitioned
adipogenesis at the onset of lipid droplet accumulation. We used
TG into lipid droplets. To rule out the possibility that FIT1 and
the 3T3-L1 adipocyte cell line, a classic adipocyte differentiation
FIT2 inhibit lipolysis of TG leading to increased lipid droplets,
cell model that produces large amounts of lipid droplets during
HEK293 cells were transfected with FIT1 or FIT2 and labeled for
differentiation of preadipocytes into adipocytes. Abundant levels of
18 h with 14C-oleic acid–BSA complex to label TG pools, and then
FIT2 mRNA and protein were detected during adipogenesis at the
TG biosynthesis was inhibited with triacsin C (21). The decrease in
onset of formation of visible lipid droplets FIT1 was not
TG levels was quantified as a measure of TG lipolysis (21).
detectable by Northern or Western blot analysis in 3T3-L1 adipo-
Expression of ADRP in cells served as a positive control for
cytes (data not shown), consistent with findings from mouse
inhibition of lipolysis (6). Lipolysis of TG was similar in cells
adipose tissues (Fig. 2 A and B). Therefore, we hypothesized that
expressing FIT1 or FIT2, compared with control HEK293 cells
if FIT2 is indeed essential for droplet formation, then suppression
and moderately reduced in cells expressing ADRP, indi-
of FIT2 expression should abolish the accumulation of lipid drop-
cating that FIT1 and FIT2 do not increase lipid droplet accumu-
lets. Preadipocytes infected with lentivirus expressing one of three
lation by inhibiting lipolysis of TG
individual shRNAs against FIT2 (FIT2shRNA1 to 3), a nonspecific
To determine whether expression of FIT proteins leads to lipid
control shRNA, or no virus were induced to differentiate into
droplet accumulation in vivo, an adenovirus-expressing FIT2 or a
adipocytes. FIT2 mRNA and protein was significantly suppressed
control adenovirus was injected into mice. At 7 days after injection,
in adipocytes infected with lentivirus-expressing FIT2 shRNA (Fig.
livers were examined for lipid droplets and TG was quantified. We
5 A and B). Examination of these adipocytes for lipid droplets
focused on FIT2 because FIT2 protein is more abundant than FIT1
showed that cells having suppressed FIT2 expression had a dramatic
Kadereit et al.
Expression of FIT2 in mouse liver. (A) Mice were injected with control
adenovirus (Adempty) or adenovirus-expressing FIT2 (AdFIT2). AdFIT2 mice
after 7 days had increased hepatic lipid droplets as judged by both H&E
staining showing cleared spaces that indicated the presence of lipid droplets
within hepatocytes (examples indicated with arrows) and Oil red O staining,indicating that these cleared spaces are TG-rich lipid droplets. These images
Lipid droplet formation induced by FIT1 and FIT2. (A) HEK293 cells
are representative of observations made on six mice per group. (Scale bar: 12
were transiently transfected with mouse FIT1, FIT2, or DGAT1, and lipid
m.) (B) TG, but not cholesterol, was significantly increased in livers of mice
droplets were visualized by using confocal fluorescence microscopy by stain-
expressing FIT2 (n ⫽ 6). Data represented as mean ⫾ SD. *, AdEmpty versus
ing with BODIPY 493/503. (Scale bar: 5 m.) (B) TG mass measurements from
AdFIT2 (P ⬍ 0.001). The large range in TG values correlated with the range in
transiently transfected HEK293 cells with the indicated constructs. Data are
FIT2 expression shown in C. (C) Western blot analysis of FIT2 expression in livers
represented as the mean ⫾ SD. *, Mock versus FIT1 or FIT2 (P ⬍ 0.001) (n ⫽ 4
of mice, indicating increased expression in AdFIT2-injected mice. -actin
transfections per construct; four independent experiments). (C) TG biosynthe-
served as a loading control.
sis was determined in transfected HEK293 cells for the indicated times. DGATs
served as positive controls for TG biosynthesis. Data are represented as the
mean ⫾ SD. *, Mock versus DGAT1 or DGAT2 (P ⬍ 0.0001) (n ⫽ 4 transfections
Knockdown of FIT2 in Zebrafish. To extend these findings to a
per construct; three independent experiments). (D) Newly synthesized TG in
whole-animal model, we turned to the zebrafish model system. The
the buoyant lipid droplet fractions from FIT1- and FIT2-expressing HEK293cells was significantly enriched compared with mock-transfected cells. Data
zebrafish has orthologs to FIT1 and FIT2 (Fig. 1B) and, as a
are represented as the mean ⫾ SD of the percentage of mock-transfected
vertebrate, has conserved lipid metabolic pathways, compared with
(control) cells. Control versus FIT1 or FIT2 (P ⫽ 0.0001) (n ⫽ 4 per transfection;
mammals (24), and transient gene knockdowns in embryos can be
two independent experiments).
generated by using morpholino antisense oligonucleotides. Similarto mouse and human FIT2 expression patterns, expressed sequencetags for zebrafish FIT2 are ubiquitously found in tissues. Zebrafish
reduction of lipid droplets and a significant reduction in total
larvae have low levels of TG and must therefore be fed a high-fat
cellular TG (Fig. 5 C and D). Because these cells have suppressed
diet to induce lipid droplet accumulation, which occurs primarily in
FIT2 expression and not a complete deficiency, the remaining
liver and intestine (25). We first tested the effect of FIT2 knock-
amount of lipid droplets in FIT2 knockdown adipocytes may be
down by using a morpholino that targets the ATG codon
because of residual amounts of FIT2 (Fig. 5 A and B). Perilipin
(FIT2morph1) and is expected to inhibit translation of FIT2.
levels strongly correlate with lipid droplet numbers and TG levels
Embryos were injected at the one-cell stage with up to 4 ng of
in adipocytes (14, 22, 23). As expected, Perilipin was reduced in all
FIT2morph1, which was well tolerated and did not cause any overt
FIT2shRNA-expressing adipocytes (Fig. 5B). We next sought to
obvious phenotype. To determine whether FIT2 knockdown in
determine whether the knockdown of FIT2 in adipocytes affected
zebrafish results in decreased lipid droplet accumulation in liver and
the process of differentiation and TG biosynthesis. Examination of
intestine in vivo, free-swimming 6-day-old larvae derived from
FIT2-suppressed cells showed that the expression of PPAR␥, a
morphant or control embryos were fed a high-fat diet for 6 h, fixed,
nuclear hormone receptor essential for adipocyte differentiation,
and compared for lipid droplet accumulation by using Oil red O
was similar in the FIT2shRNA2 and FIT2shRNA3 adipocytes,
staining. The intestine, liver, and swim bladder of control and
compared with controls, with moderate variation in levels between
control morpholino zebrafish stained red for TGs after 6 h of
samples, but was decreased in FIT2shRNA1 adipocytes, which had
high-fat feeding (Fig. 6A), which was similar to previously published
the most reduced levels of FIT2 and TG. Expression of the PPAR␥
data (25). The FIT2 morphant fish showed a near absence of
target genes, aP2, and adiponectin/ACRP30 followed a similar
intestinal and hepatic lipid droplet staining, although feeding
trend, with decreased levels in the FIT2shRNA1 adipocytes
behavior was normal as determined by ingestion of nonabsorbable
(Fig. 5A).
fluorescent microbeads (Fig. 6A). In the FIT2 morphant fish,
The decrease in TG levels in FIT2shRNA cells could be because
whole-body Oil red O staining as a measurement of TG levels was
of decreased TG biosynthesis. To test this possibility, the rate of TG
significantly decreased, compared with controls (Fig. 6B). To
biosynthesis was examined in FIT2shRNA and control cells 2, 4, 6,
determine that reduced lipid droplets and TG are specifically
and 9 days after induction of differentiation. TG biosynthesis was
because of morpholino-directed knockdown of FIT2, we compared
significantly decreased in FIT2shRNA cells (Fig. 5E). In contrast,
the effects of a second morpholino that targets the 5⬘ splice site of
TG lipase activity was similar in FIT2shRNA cells, compared with
FIT2 mRNA (FIT2morph2). When injected into embryos,
controls In summary, the data indicate that FIT2
FIT2morph2 significantly inhibits splicing of its single intron, re-
knockdown in 3T3-L1 adipocytes resulted in decreased lipid droplet
sulting in the introduction of four in-frame stop codons
accumulation and significantly decreased TG biosynthesis.
and is therefore predicted to abolish FIT2 expression. Indeed,
Kadereit et al.
PNAS 兩 January 8, 2008 兩 vol. 105 兩 no. 1 兩 97
Morpholino-mediated knockdown of FIT2 in zebrafish. (A) FIT2
morphants showed decreased staining for Oil red O in liver (outlined by
dashed line in wild type) and intestine (outlined by solid line in wild type),
compared with control fish (no morpholino control, WT; and nonspecific
control morpholino, contmorph). Swim bladders of zebrafish stain with Oil
red O (indicated by arrow). FIT2 morphants did not exhibit defects in feedingas judged by ingestion of nonabsorbable fluorescent microbeads. A WT
control fed a high-fat diet without microbeads is shown (no beads) to dem-
onstrate that fluorescence is because of ingested microbeads, not autofluo-
rescence of fish. These images are representative of n ⫽ 400 fish, three
independent experiments. (Scale bar: 0.4 mm.) (B) Quantification of Oil red O
staining by spectrophotometric analysis in WT, contmorph, FIT2morph1, and
FIT2morph2 fish (n ⫽ 20 fish per group, averaged over four independent
experiments). *, WT versus morphants (P ⬍ 0.001) and shown as mean ⫾ SD.
proteins are evolutionarily conserved and exclusively located in the
ER, the site of TG biosynthesis and incidentally the long-proposed
site of lipid droplet biogenesis; (ii) overexpression of FIT proteins
Days of differentiation
in cultured cells or in mouse liver in vivo results in the accumulation
shRNA-mediated knockdown of FIT2 in adipocytes. 3T3-L1 cells were
of TG-rich lipid droplets; (iii) unlike DGATs, FIT proteins do not
not infected or were infected with lentivirus-expressing shRNA sequences
mediate the biosynthesis of TG, but enhance the partitioning of TG
targeting murine FIT2 (FIT2shRNA1,2,3) or control shRNA (contshRNA) and
into lipid droplets, placing FIT proteins functionally downstream of
differentiated for 7 days. (A) Northern blot analysis shows that FIT2shRNAs
DGATs; and (iv) shRNA-mediated depletion of FIT2 in 3T3-L1
significantly reduced FIT2 mRNA levels compared with noninfected control
adipocytes or knockdown of FIT2 in zebrafish embryos dramati-
(cont) and contshRNA-infected cells. Expression of adipocyte differentiation
cally reduced the accumulation of lipid droplets. Curiously, the
markers PPAR␥, aP2, and adiponectin/ACRP30 are shown. The ethidium bro-mide-stained RNA gel serves to indicate loading. (B) Western blot analysis
yeast FIT2 ortholog FIT2b was identified 13 years ago in a genetic
shows that FIT2 and Perilipin were reduced in FIT2shRNA cells, compared with
screen as one of many genes when mutated resulted in myo-inositol
controls. (C) FIT2shRNAs reduced lipid droplet accumulation in differentiated
auxotrophy in the presence of choline and named SCS3 (26).
3T3-L1 cells as visualized by BODIPY493/503 staining of lipid droplets. (Scale
Hosaka and coworkers (26) speculated that SCS3 is involved in
bar: 10 m.) (D) Quantification of cellular TG in differentiated 3T3-L1 cells
regulating inositol synthesis, but did not quantify levels of inositol
shows reduced TG levels in the FIT2 knockdown cells. Data represented as
phospholipids in SCS3 mutants or provide evidence that SCS3
mean ⫾ SD. *, FIT2shRNA1–3 versus controls P ⬍ 0.0001. (E) TG synthesis
regulates this pathway (26). Thus, a role of SCS3 in regulating
measurements were performed at the indicated time points during differen-
inositol phospholipid biosynthesis is purely speculative. Moreover,
tiation. Each time point represents three independent samples for each timepoint and is shown as mean
FIT2a was not identified from this genetic screen. Since this original
⫾ SD. *, FIT2shRNA2,3 versus controls (P ⬍ 0.001).
A–D are representative of three independent experiments. E is representative
publication of SCS3, no further studies on SCS3 (FIT2b) have been
of two independent experiments.
reported, but the proteins involved in regulating phospholipidbiosynthesis in S. cerevisiae have been identified and SCS3 is notamong them (27, 28). In our studies, we have not found changes in
treatment with FIT2morph2 resulted in a similar decrease in lipid
levels of phospholipids, including inositol phospholipids in HEK293
droplets in the liver and intestine and whole-body Oil red O
cells overexpressing FIT1 or FIT2
staining, compared with FIT2morph1 (Fig. 6 A and B). Moreover,
We initially identified FIT genes as cDNAs up-regulated by
overexpression of zebrafish FIT2 in HEK293 cells confirmed that
PPAR␣ in liver. On the one hand, this finding is somewhat
zebrafish FIT2 functions to produce lipid droplets and increase TG
perplexing given that activation of PPAR␣ is associated with
levels in mammalian cells further indicating
increased expression of genes involved in the oxidation of fatty acids
conservation of function. Together these data show that FIT2 is
(16). On the other hand, activation of PPAR␣ in the liver also
important for lipid droplet accumulation in mouse adipocytes and
up-regulates genes involved in lipogenesis, including malic enzyme,
during embryogenesis in zebrafish liver and intestine.
acetyl-coA carboxylase, fatty acid synthase, steroyl-CoA desaturase1, glycerol-3-phosphate acyltransferase (29, 30), and two lipid
droplet-associated proteins that are part of the PAT family, PAT-
The present study describes a highly conserved family of proteins
1/MLDP/OXPAT/LSDP5 (31, 32) and ADRP (33), indicating that
that are important for the accumulation of lipid droplets. Four
liver PPAR␣ regulates both lipid catabolic and anabolic pathways.
major lines of evidence support our conclusion that FIT proteins
Notably, FIT2 expression is up-regulated during 3T3-L1 differen-
are important for the accumulation of lipid droplets: (i) FIT
tiation similarly as PPAR␥ at a time when lipid droplets are known
Kadereit et al.
to accumulate, giving 3T3-L1 cells their adipocyte phenotype.
proteins should facilitate the development of reagents to regulate
Indeed, we found that FIT2 expression is up-regulated by the
FIT expression or activity to treat diseases associated with excessive
treatment of 3T3-L1 cells with rosiglitazone, a specific PPAR␥
lipid droplet accumulation, such as obesity, type 2 diabetes, and
agonist, further supporting the notion that FIT2 is additionally
regulated by PPAR␥ (B.K., D.M., and D.L.S., unpublished data).
Materials and Methods
shRNA-mediated knockdown of FIT2 in 3T3-L1 adipocytes
resulted in a dramatic decrease in lipid droplets and TG levels.
Reagents. Rabbit polyclonal antibodies were raised against peptides corre-
sponding to the C-terminal 15 amino acids of murine FIT1 and FIT2 and
Interestingly, this decrease, with the exception of FIT2shRNA1
validated by using lysates from HEK293 cells expressing murine FIT1 and
adipocytes, was not associated with a significant decrease in PPAR␥
and the PPAR␥ target genes aP2 or adiponectin/ACRP30, indi-cating that FIT2-depleted adipocytes differentiate similarly to
Membrane Fractionations and Isolation of Lipid Droplets. See
control cells. This finding suggests that the primary effect of FIT2
for details.
knockdown is on TG lipid droplet accumulation and not ondifferentiation. Interestingly, the decrease in TG levels in FIT2
TLC Assays and TG and Cholesterol Measurements. TG or cholesterol quantified
by enzymatic assay (Infinity triglyceride kit and Infinity cholesterol; Thermo-
knockdown adipocytes was associated with decreased TG biosyn-
Electron). See for details.
thesis. This finding is in contrast to our findings that overexpressionof FITs in HEK293 cells does not enhance TG biosynthesis. One
Confocal Immunofluorescence Microscopy. HEK293 cells were transiently trans-
possible explanation for these findings is that an inhibition in the
fected with expression plasmids for mFIT1-V5, mFIT2-V5, or RFP with the
ability to produce lipid droplets, and therefore store TG in FIT2
ER-retention signal KDEL or with the live cell Gogli marker protein galactosyl-
knockdown adipocytes, results in a partial feedback inhibition in
transferase-Tag-RFP (35, 36) and processed for confocal microscopy as de-scribed in
TG biosynthesis without an entire blockade in adipogenesis. Thisscenario would limit the buildup of lipotoxic lipid intermediates
TG Hydrolysis and Biosynthesis Assays. For a detailed description see
from the glycerol-3-phosphate pathway, such as diacylglycerol (34).
Nonetheless, knocking down FIT2 in zebrafish resulted in de-creased neutral lipid accumulation in liver and intestine, providing
Lentivirus shRNA. A detailed description of shRNA sequences and lentivirus
confirmatory in vivo evidence in a whole-animal vertebrate model
production can be found in 3T3-L1 preadipocytes
that FIT2 is important for lipid droplet accumulation. Moreover,
were infected with a MOI of 10 –50 and selected for 7 days with blasticidin.
Cells were consequently plated and differentiated for 7 days to produce
overexpression of FIT2 in mouse liver in vivo increased levels of
mature adipocytes. shRNAi experiments were repeated two independent
TG-rich lipid droplets. Presently, the mechanism by which FIT
times for FIT2shRNA1 or three independent times for FIT2shRNA2 and
proteins mediate lipid droplet accumulation is not known. We have
found no evidence by using immunoprecipitation techniques thatFIT proteins interact with the ubiquitously expressed lipid droplet-
Morpholino Knockdown of FIT2 in Zebrafish. Sequences of morpholinos and
associated protein ADRP (data not shown) that plays a role in lipid
experimental details can be found in
droplet metabolism.
ACKNOWLEDGMENTS. This work was supported in part by National Institutes
The sum of our data support the conclusion that FIT proteins are
of Health Grants P30 DK41296 (through the Marion Bessin Liver Research
necessary for lipid droplet accumulation. The identification of FIT
Center) (to D.L.S.) and R01 HL064282 (to T.E.).
1. Martin S, Parton RG (2006) Nat Rev Mol Cell Biol 7:373–378.
21. Brasaemle DL, Rubin B, Harten IA, Gruia-Gray J, Kimmel AR, Londos C (2000) J Biol Chem
2. Spiegelman BM, Flier JS (2001) Cell 104:531–543.
275:38486 –38493.
3. Brasaemle DL, Dolios G, Shapiro L, Wang R (2004) J Biol Chem 279:46835– 46842.
22. Xu G, Sztalryd C, Londos C (2006) Biochim Biophys Acta 1761:83–90.
4. Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG (2004) J Biol Chem 279:3787–3792.
23. Brasaemle DL, Barber T, Kimmel AR, Londos C (1997) J Biol Chem 272:9378 –9387.
5. Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, Kuhnlein RP (2006) Mol
24. Tocher D (1995) in Biochemistry and Molecular Biology of Fishes, eds P Hochachka, T
Cell Proteomics 5:1082–1094.
Mommsen (Elsevier, New York), pp 119 –157.
6. Chang BH, Chan L (2007) Am J Physiol 292:G1465–G1468.
25. Schlegel A, Stainier DY (2006) Biochemistry 45:15179 –15187.
7. Mullner H, Daum G (2004) Acta Biochim Pol 51:323–347.
26. Hosaka K, Nikawa J, Kodaki T, Ishizu H, Yamashita S (1994) J Biochem (Tokyo)
8. Zechner R, Strauss JG, Haemmerle G, Lass A, Zimmermann R (2005) Curr Opin Lipidol
27. Chen M, Hancock LC, Lopes JM (2007) Biochim Biophys Acta 1771:310 –321.
9. Londos C, Brasaemle DL, Schultz CJ, Segrest JP, Kimmel AR (1999) Semin Cell Dev Biol
28. Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP (2004)
Science 304:1644 –1647.
10. Duncan RE, Ahmadian M, Jaworski K, Sarkadi-Nagy E, Sul HS (2007) Annu Rev Nutr 27: 79–101.
29. Castelein H, Gulick T, Declercq PE, Mannaerts GP, Moore DD, Baes MI (1994) J Biol Chem
11. Cases S, Smith SJ, Zheng YW, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB,
269:26754 –26758.
Lusis AJ, et al. (1998) Proc Natl Acad Sci USA 95:13018 –13023.
30. Knight BL, Hebbachi A, Hauton D, Brown AM, Wiggins D, Patel DD, Gibbons GF (2005)
12. Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, Farese RV, Jr. (2001)
Biochem J 389:413– 421.
J Biol Chem 276:38870 –38876.
31. Wolins NE, Quaynor BK, Skinner JR, Tzekov A, Croce MA, Gropler MC, Varma V,
13. Novikoff AB, Novikoff PM, Rosen OM, Rubin CS (1980) J Cell Biol 87:180 –196.
Yao-Borengasser A, Rasouli N, Kern PA, et al. (2006) Diabetes 55:3418 –3428.
14. Wolins NE, Brasaemle DL, Bickel PE (2006) FEBS Lett 580:5484 –5491.
15. Robenek H, Hofnagel O, Buers I, Robenek MJ, Troyer D, Severs NJ (2006) J Cell Sci
32. Yamaguchi T, Matsushita S, Motojima K, Hirose F, Osumi T (2006) J Biol Chem
119:4215– 4224.
16. Staels B, Dallongeville J, Auwerx J, Schoonjans K, Leitersdorf E, Fruchart JC (1998)
33. Dalen KT, Ulven SM, Arntsen BM, Solaas K, Nebb HI (2006) J Lipid Res 47:931–
Circulation 98:2088 –2093.
17. Wolfe KH, Shields DC (1997) Nature 387:708 –713.
34. Schaffer JE (2003) Curr Opin Lipidol 14:281–287.
18. Almsherqi ZA, Kohlwein SD, Deng Y (2006) J Cell Biol 173:839 – 844.
35. Cole NB, Smith CL, Sciaky N, Terasaki M, Edidin M, Lippincott-Schwartz J (1996) Science
19. Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N,
273:797– 801.
Lippincott-Schwartz J (2003) J Cell Biol 163:257–269.
36. Merzlyak EM, Goedhart J, Shcherbo D, Bulina ME, Shcheglov AS, Fradkov AF,
20. Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003)
Gaintzeva A, Lukyanov KA, Lukyanov S, Gadella TW, et al. (2007) Nat Methods
Nature 425:686 – 691.
Kadereit et al.
PNAS 兩 January 8, 2008 兩 vol. 105 兩 no. 1 兩 99
Source: http://prodeinsteinwebsite.vipprod.dmz.yu.edu/labs/erik-snapp/publications/Kadereit2008PNAS.pdf
Your 2015 Prescription Drug List effective January 1, 2015 Please read: This document contains information about commonly prescribed medications. For additional information: Call the toll-free member phone number on the back of your health plan ID card. Visit myuhc.com® • Locate a participating retail pharmacy by ZIP code.
Concentrations de médicaments, d'hormones et de quelques autres contaminants d'intérêt émergent dans le Saint-Laurent et dans trois de ses tributaires Problématique Une part importante des produits pharmaceutiques et de soins Au Québec comme ailleurs, des échantillonnages réalisés dans personnels (PPSP) que nous utilisons couramment, comme les dernières décennies ont permis de déceler la présence de les crèmes hydratantes pour la peau, les shampoings et les nouveaux contaminants dans les cours d'eau et d'y constater dentifrices, se transfère dans l'eau au moment de la douche de nouveaux effets sur les organismes aquatiques. Par exemple, ou d'autres pratiques d'hygiène. D'autres produits, comme les dans le fleuve Saint-Laurent en aval de Montréal, on a constaté médicaments pris par voie orale, sont en grande partie éliminés que des poissons et des moules d'eau douce se féminisaient dans les excrétions humaines et se retrouvent eux aussi dans (Aravindakshan et al., 2003; Blaise et al., 2003). Ce phénomène les eaux usées domestiques. Les industries où sont fabriquées est attribuable à la présence dans l'eau d'hormones naturelles ces substances et la disposition inappropriée des produits ou de synthèse, ou de produits chimiques pouvant agir comme non utilisés (ex. : médicaments jetés dans les toilettes) sont des hormones, c'est-à-dire des « perturbateurs endocriniens ». d'autres sources de PPSP dans les eaux usées industrielles et municipales.











