Viagra gibt es mittlerweile nicht nur als Original, sondern auch in Form von Generika. Diese enthalten denselben Wirkstoff Sildenafil. Patienten suchen deshalb nach viagra generika schweiz, um ein günstigeres Präparat zu finden. Unterschiede bestehen oft nur in Verpackung und Preis.
Type of article: original
Molecular and Biochemical Diagnosis (MBD)
Vol 1, No 2, 2014
Original Article
In Silico Studies on Fingolimod and Cladribine Binding to p53
Gene and Its Implication in Prediction of Their Carcinogenicity
Potential
Karim Mahnam1, Azadeh Hoghoughi1
1. Biology Department, Faculty of Science, Shahrekord University, Shahrekord, Iran
Abstract
Background: New drugs namely; cladribine and fingolimodare known to be effective in
treatment of multiple sclerosis (MS). The interaction of these drugs with the promoter region
of the p53 gene may alter p53role in cancer progression. The aim of this study was to known
the interaction of these compounds with p53 gene.
Methods: Binding free energy of the cladribine, fingolimod and their modified drugs for the
p53 gene promoter were investigated using docking, 100 ns molecular dynamics simulations
and MM/PBSA calculation.
Results: The results showed that both cladribine and modified cladribine (replacing -OH on
carbon 3´ ribose sugar with -CH3 group) can bind the minor groove of p53 promoter, and
inhibit the binding of transcription factors and expressio n of p53. However, fingolimodand its
derivatives showed relatively weaker interaction with p53 promoter
Conclusions: Based on in silico studies we showed that the binding of cladribine to the p53
gene is stronger than that of fingolimod, hence it seems tha t the former drug can pose
potential carcinogenic effects. The binding power and carcinogenic effect of sm-fingolimod
(removing four carbons from its aliphatic tail) is more than that of fm-fingolimod (removing
one carbon from its aliphatic tail).
Keywords: Cladribine, Fingolimod, Molecular dynamics simulation, MM/PBSA, p53 gene
deficits, followed by progressive neurological
Multiple sclerosis (MS) is a demyelinating
deteriorations (Navikas
et al., 1996).
inflammatory disorder of the central nervous
The first oral disease-modifying drug approved
system (CNS) with autoimmune responses.
food and drug administration (FDA) is
The degree of axonal destruction is variable
Fingolimod (Gilenya, Novartis) (Fig. 1A) to
(Calabresi, 2004). The route of MS is highly
postpone progression of physical disability in
varied and unpredictable so that it may be
sphingosine kinase to the active metabolite;
fingolimod phosphate, which in turn blocks
*
C orre sponding author. Karim Mahnam, PhD.
migration of lymphocytes from lymphnodes,
Biology Department, Faculty of Science, Shahrekord University, P.O. Box: 8818634141,Shahrekord, Iran
thereby reducing the number of lymphocytes
T el., +98-038-4424402; Fax, +98-38-4424419 Email:
[email protected]
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
K. Mahnam et al.
in peripheral blood (Cohen
et al., 2007). The
incorporated into DNA, thereby causes down-
possible mechanism of the therapeutic effect of
regulation of cellular ribonucleotidereductase
fingolimod in MS is through the reduction of
and inhibit DNA synthesis (Foley
et al., 2004).
TATA element of the promoter is recognized
(Francesca, 2007).
by TATA binding protein (TBP). Foley
et al
showed that positions in the TATA sequence
are most severely affected by cladribine
incorporation (Foley
et al., 2004).
In general, drug targets are cytoplasmic
proteins, membrane receptors or membrane-
bound proteins, nuclear proteins, DNA etc.
Small aromatic compounds can bind DNA by
A: Covalent bond; through their functional
groups irreversibly attached to DNA, leading
to inhibition of DNA synthesis processes and
Figure 1. Structure of fingolimod (A) and cladribine (B).
cell death such as Cisplatin and Mitomycin
Fingolimod has been associated with reduce
(Elizondo-Riojas
et al., 2001).
heart rate (bradycardia) and usually fatal
B: Non-covalent bond; by intercalation (such
infections such as cancer (Cohen
et al., 2007).
Another drug used to trea
intominor groove binding such as; Netropsin,
(HCL, leukemic reticuloendotheliosis) and
Distamycin and into major groove binding;
is cladribine (Leustatin, Litak and Movectro™)
such as Norfloxacin (Neidle
et al., 1987).
The tumor suppressor
p53 gene as an
important tumor suppressor gene continually is
transcribed to prevent cancer.
P53 gene is the
ibine). It is a and acts as
most frequently mutated gene in human tumors
suppressor of the immune system. Possible
(Vogelstein
et al., 2010). In some cancers,
side effects of the cladribine include fever,
transcription of the p53 gene is reduced (Bai
et
infection, anemia and cancer. CldAdo is taken
al., 2006).
up by cells, converted to 2-chloro-2´-deoxy
The Molecular Mechanics/Poisson–Boltzmann
Surface Area (MM-PBSA) method has been
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
In silico fingolimod and cladribine binding to p53 gene
used to calculate relative free energies of DAPI
sequences of DNA (Spacková
et al., 2003).
Currently the computational techniques are
Promoter of
p53 gene has 52 pair nucleotides. The
widely applied in chemistry and biology
sequence of the 5´ to 3´ strand of promoter of
p53
ranging from the quantum mechanics of
gene that was applied for this study was 5´-
molecules to the dynamics of large complex
molecular aggregates. Molecular interactions
steer chemical reactions, phase transitions and
(Reisman
et al., 1993). 3D structure of
p53
other physical phenomena and can be studied
promoter was generated via 3D-Dart (3DNA-
via molecular dynamics (MD) simulations,
Driven DNA Analysis and Rebuilding Tools)
showing the detailed motion of molecules or
(haddock.science.uu.nl/
atoms as a function of time. The MD
services/3DDART). Also, geometries of all
simulations provide powerful links between
ligands were obtained from Arguslab software
the model equilibrium, minimal geometries of
proteins and DNA and binding free energy of
Lab.html) via molecular mechanics methods
drugs (Karplus
et al., 2005). The calculation of
under MM+ force fields and used for docking
relative binding free energies of ligands to a
and MD simulation studies. The atomic
receptor has been used for better understanding
charges of all ligands were calculated with the
of molecular interactions of proteins with
Merz−Kol man electrostatic potential fitting
procedure in the Gaussian quantum chemistry
et al., 2005).
package (Frisch
et al., 1998). This was
In our ongoing project, we have performed
performed by means of a Hartee-Fock wave
some theoretical studies to investigate the
function obtained in a 6-31G* basis set for
mechanism of binding of cladribine and
compatibility with the partial charges from the
fingolimodto promoter of
p53 gene. In
AMBER force field that was used for
p53
addition, the effect of some structural
modifications of these drugs in binding their
free energy to promoter of
p53 gene has been
calculation was done using this command:
HF/6-31G* Pop=MK IOp (6/33=2, 6/41=10,
6/42=17)
et al., 2011). Cladribine was
underlying carcinogenicity of cladribine or
modified by replacing OH on carbon 3´ ribose
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
K. Mahnam et al.
sugar with CH3 group. Modification of
fingolimod was done by removing one carbon
2. Molecular dynamic simulations
fingolimod) from the aliphatic hydrocarbon
Five molecular dynamics simulation of ligands
tails. All images were generated with
complexes with
p53 promoter sequence were
performed. The cycle time for each simulation
was 20 ns. Then, one hundred ns MD
Theoretical studies were done in three
simulations were applied. MD simulation and
following sections:
molecular mechanic (MM) minimization were
performed using GROMACS 4.5.3 package
1. Docking
under Amber99 force fields (Van der Spoel
et
Autodock 4 software was used for docking
al., 2005; Berendsen
et al., 1995; Hess
et al.,
studies (Morris
et al., 1998). The grid box size
2008 and Lindahl
et al., 2001). Topologies of
was set at 9090118 Å and spacing between
ligands were generated by acpype/Antechamber
grid points 0.375 angstrom. The
p53 promoter
based on a General Amber Force Field (GAFF)
structures were fixed during docking, while the
(Sousa
et al., 2012). MD simulations were
drugs were flexible. Grid searching was
carried out in an NPT ensemble with periodic
performed by a local search genetic algorithm
boundary conditions. Van der Waals forces
(LGA) to locate the ligands in the lowest
were treated using a cut-off of 12 Å. The
binding energy. Routine procedures and
electrostatic interactions were calculated using
default parameters were used in the docking
the Particle-Mesh Ewald model with a 14 Å
except dstep, tstep and qstep that were
cut-off (Darden
et al., 1993).The complexes
considered 0.5 Å, 0.5°, 5° respectively
were solvated by a layer of water of at least 12
(Majumdar
et al., 2011).
Å in all directions. The frequency to update the
All ligands (cladribine, modified cladribine,
neighbor list was 10 ps. MD simulation was
accomplished in four steps for each system. In
fingolimod) were docked on
p53 promoter.
the first step, the entire system was minimized
Two hundred docking runs were performed for
using the steepest descent followed by
each docking. The best pose with the lowest
conjugate gradient algorithms. In the second
binding energy and the most populated
step, the solvent and Na+ ions were allowed to
conformation in each cluster was chosen as the
evolve using minimization and molecular
initial structure in the molecular dynamics
dynamics in the NVT ensemble for 500 ps and
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
In silico fingolimod and cladribine binding to p53 gene
in the NPT ensemble for 1000 ps at 100 K,
energy of a DNA molecule to a ligand
where the initial configuration of the structures
molecule in a solution can be defined as:
was kept fixed. In the third step, in order to
∆Gbinding=Gcomplex-(GDNA+Gligand) Eq.1
obtain equilibrium geometry at 300 K and 1
"A MD simulation is performed to generate a
atm, the system was heated at a weak
thermodynamically weighted ensemble of
temperature coupling (τ = 0.1 ps) and pressure
structures" (Kumari
et al., 2014). The free
coupling (τ = 0.5 ps). The Berendsenalgorithm
energy term is calculated as an average over
was chosen for thermostat and barostat in
the considered structures:
equilibration phase (Berendsen
et al., 1984). To
constrain the lengths of hydrogen-containing
Total molecular mechanical energies EMM is
bonds, the LINCS algorithm was used (Hess
et
calculated by using GROMACS utility with
al., 1997). The temperature of the system was
the AMBER99 force field. -T<SMM> is the
then increased from 100 K to 300 K and the
solute entropic contribution. Gsolvation represents
velocities at each step were re-accredited
the free energy of solvation and consists of two
Maxwell-Boltzmann
parts: Gpolar or GPB and nonpolar contributions,
distribution at that temperature and equilibrated
Gnonpolar. GPB is generated from the electrostatic
for 200 ps. In the final (production) step, 20 ns
MD simulations at 300 K with a time step of 2
(Massova
et al., 1999).
fs was performed for each complex and final
In the current study, Gpolar was calculated using
structures were obtained. The thermostat and
APBS (Adaptive Poisson-Boltzmann
barostat for production step were Nosé-Hoover
Solver program) method (Baker
et al., 2001)
via the non-linearized Poisson Boltzmann
(Berendsen
et al., 1984). In all simulations, two
equation. The non-polar contribution, Gnonpolar
single strands of DNA were constrained to each
was considered to be proportional to the
other (Cheatham
et al, 1998). Potential and
solvent accessible surface area (SASA).
kinetic energies and temperature at the last 5 ns
In the MM/PBSA approximation and for
were calculated using g_energy command of
estimating Gfree-DNA and Gfree-ligand, snapshots
Gromacs package. Other analyses were
collected from the MD run for the DNA-ligand
performed by using Gromacs package.
complex were used. After equilibration,
snapshots of complex, DNA and ligand
3. MM/PBSA calculation
(without water molecules) were taken every 50
As indicated by Kumari, the binding free
ps for calculating the enthalpy.
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 ( 2014), 105-122
K. Mahnam et al.
Binding free energy calculations based on the
have suggested that including corrections for
MM/PBSA approach can be performed either
changes in the configurational free energy of the
according to the three trajectories method
system lead to only a small improvement in the
(TTM) or according to the single trajectory
total. We decided to neglect the entropic term in
method (STM). In our work, MM/PBSA
our calculations. The last 5 nanosecond of the
calculations were performed according to the
MD simulations was considered for MM/PBSA
STM protocol. A single trajectory run for the
complex is required for this method, whereby
The energy components EMM, Gpolar and Gnon-
both the DNA and ligand structures are
polar of each complex were calculated for 100
extracted directly from the complex structure
snapshots extracted every 50 ps from the
(Huo
et al., 2002), thus zeroing out the Eint
production trajectories at the last 5 ns. To
term. In this case, the DNA and the ligands are
calculate Gpolar, a box was generated using the
assumed to behave similarly in the bound and
extremes coordinates of the molecular complex
in the free forms.
in each dimension. A coarse-grid box (cfac =3)
In the MM/PBSA approximation, EMM+Gsolv
was obtained when the box expanded in each
account for the enthalpy change is associated
dimension by two-fold. A finer grid-box is
with complex formation. The computational
then placed within the coarse grid-box
determination of binding free energies requires
extending 50 Å (fadd=50) from the complex's
the calculation of the entropic contributions to
extremes coordinates in each direction. An
complex formation including conformational
ionic strength of 0.6 M NaCl with radii of 0.95
changes in the rotational, translational and
and 1.81 Å, respectively for sodium and
vibrational degrees of freedom of the solute.
chloride ions was used during all Gpolar
The MM/PBSA method was used by
calculations. The values for vacuum (vdie) and
g_mmpbsa command (Baker
et al., 2001; Pronk
solvent (sdie) dielectric constants were taken
et al., 2013; Eisenhaber
et al., 1995 and Kumari
as 1 and 80 respectively. The solute (pdie)
et al., 2014). In this module, entropic terms are
dielectric constant was assigned a value of
not included and therefore it is unable to give
eight. Subsequently, the binding free energy of
the absolute binding energy. Thus, it is proper
each snapshot was calculated for each complex
to calculate the relative binding energies for
using a combination of Eq.1 and 2 without
instance, to compare different ligands binds to
entropic contributions in the binding energy
the same receptor. In addition, the net entropic
(Kumari
et al., 2014 and Brown
et al., 2009
contribution is often small, and multiple studies
and Gohlke
et al., 2004 and Kar
et al., 2011
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (Autumn 2014), 105-122
In silico fingolimod and cladribine binding to p53 gene
and Bradshaw
et al., 2011).
p53 sequence are negative; so these drugs are
able to bind the
p53 promoter. Also, binding
Results and Discussion
position of these ligands were mentioned. The
1. Docking
positions of all compounds were in the minor
Investigation of the docking results in Table 1
groove of
p53 promoter. The binding position
shows that the binding free energy of cladribine,
of cladribine and modified cladribine are 5´-
fingolimod, modified cladribine (replacing OH
T15T16G17-3´ nucleotide; and those of
on carbon 3´ ribose sugar with CH3 group), the
first and second modified fingolimod (removing
one carbon or four carbons from the aliphatic
modified fingolimod) to
p53 promoter are 5´-
hydrocarbon tail of fingolimod respectively) to
G30T31T32T33T34-3´ nucleotides.
Table 1.Van der Waals (VDW)contribution, Electrostatic contribution (Elec) and the lowest binding free energy
(L.B) of native and modified cladribine, fingolimod to p53 promoter, nucleotides15-34 are shown.
Compound
VDW + Hbond + desolvation
S equence of binding position
(kcal/mol)
(kcal/mol)
Cladribine
5´- T15T16G17-3´
Modified cladribine
5´-T15T16G17-3´
Fingolimod
5´-G30T31T32T33T34-3´
5´-G30T31T32T33T34-3´
S m-fingolimod2
5´-G30T31T32T33T34-3´
1. First modification of fingolimod (i.e. deleting one carbon of fingolimod tail). 2. Second modification of fingolimod (i.e. deleting four carbon of fingolimod tail).
These sequences are the positions of binding of
fingolimod are weaker to bind
p53 promoter.
transcription factors such as USF (upstream
In all cases, Van der Waals (plus Hbond and
stimulatory factor) or TFE3 (transcription
desolvation) contributions are more negative
factor E3) (Kim
et al., 2008; Yasumoto
et al.,
important than electrostatics
1994). Binding free energy of modified
interactions (Table 1).
cladribine to
p53 promoter is lower than that
for cladribine, it means that the binding of
2. Molecular dynamics simulation
modified cladribine is stronger than that for
Table 2 shows the results of average potential
cladribine but binding free energy of the first
and kinetic energies, temperature, root mean
and second modified fingolimod to
p53
square deviation (RMSD) of
p53 promoter and
promoter are more than that for fingolimod, it
ligands RMSD relative to initial positions
means that the first and the second modified
during the last 5 ns of 20 ns MD simulation.
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
K. Mahnam et al.
There are small variations in potential and
sufficient and stable under the simulation
kinetic energy, temperature and RMSD of the
conditions and thermal equilibrium of the
p53 promoter during the last 5 ns of MD
systems. By investigating the final structures
simulation with a very low ratio of the total
of 20 ns MD simulation it appeared that the
energy drift to the average total energy (Table
two strands of the
p53 promoter remained
3). This shows that the simulations were
together during 20 ns simulations.
Table 2. The potential energy (P), kinetic energy (K) and temperature (T) and radius of gyration (Rg) and
RMSD of
p53 promoter and drugs at complex during the last 5 ns of MD simulations.
RMS D of p53
Promoter at
promoter at
P (kcal/mol)
K (kcal/mol)
Cladribine
Modified cladribine
Fingolimod
S m-fingolimod2
1. Fm-Fingolimod:First modification of fingolimod (i.e. deleting one carbon of fingolimod tail); 2. Sm-Fingolimod: Second modification of fingolimod (i.e. deleting four carbon of fingolimod tail). *. Nanometer
Table 3: The ratio of the total energy drift to average of total energy during 20 ns MD simulations of all species.
Ratio of the total energy drift to
S ystem name
average of total energy (
10-5)
Cladribine
Modified cladribine
Fingolimod
S m-fingolimod2
1. Fm-Fingolimod: First modification of fingolimod (i.e. deleting one carbon of fingolimod tail) 2. Sm-Fingolimod: Second modification of fingolimod (i.e. deleting four carbon of fingolimod tail).
Also, small RMSDs of ligand atoms during
neighbors if the backbone RMSD between
simulation relative to the starting position
them was less than 0.2 nm.
(Table 2) showed that the ligands reach to
The middle structure of the most populated
stable positions.
structures obtained from clustering of trajectories
To determine the relative populations of all
during the last 5 ns of MD simulation showed
conformations, the trajectories were clustered
that cladribine and modified cladribine stay in
using g_cluster command of the Gromacs
the minor groove of
p53 promoter in 5´-
package. Two conformations were considered
T16G17A18-3´ sequence however, fingolimod,
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
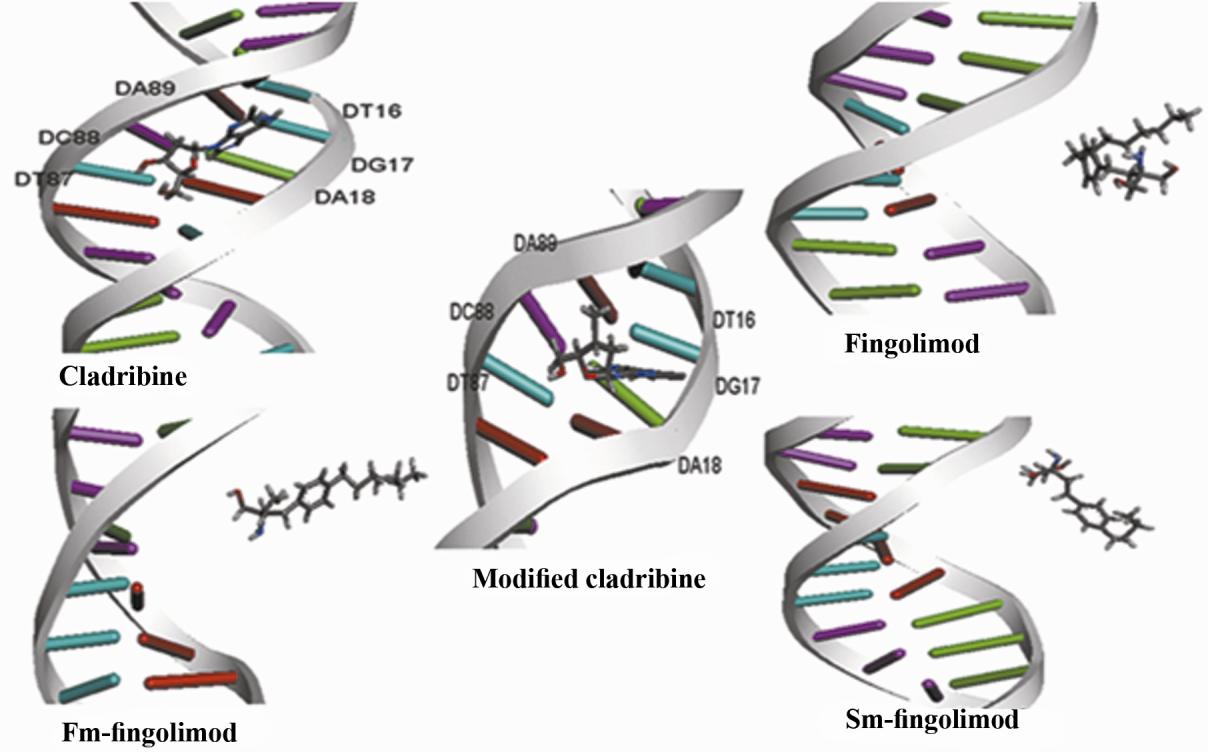
In silico fingolimod and cladribine binding to p53 gene
the first and second modified fingolimod go
away from their initial docking positions (Fig. 2).
Figure2.The middle structure of the most populated structures of drugs -DNA complex during the last 5 ns MD
simulation.The number of the nucleotides in double strandedp53 promoter was mentioned in Table 4. fm-
fingolimod: First modification of fingolimod,i.e. deleting one carbon of fingolimod tail. sm-fingolimod: Second
modification of fingolimod, i.e. deleting four carbon of fingolimod tail.
The number of nucleotides in double-stranded
fingolimod (belongs to 17.1 ns) and second
p53 promoter has been indicated in Table 4. In
modified fingolimod (belongs to 16.98 ns), no
the middle structure of the most populated
hydrogen bonds seen with p53 promoter.
structures of cladribine (belongs to 19.6 ns)
The average solvent accessible surface area
and modified cladribine (belongs to 18.68 ns)
(SASA) of the ligand atoms during the 20 ns MD
in complex with p53 promoter, guanosine 17
simulation were calculated by g_sas command
(H22 and N3 and O4´ atoms) and adenosine 89
and non-hydrogen atoms with SASA less than 10
(N3 atom) of double stranded p53 promoter,
Å2 were determined. These atoms probably bind
have hydrogen bonds with cladribine. In
to the p53 promoter during MD simulation. The
middle structure of the most populated
results showed that cladribine bind the p53
structures of MD simulation of fingolimod
promoter via its N2, N4, O1, C3, C2 and N3
atoms (these atoms were shown in Fig. 1A).
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
K. Mahnam et al.
Table 4. The frequency of nucleotides in double-stranded p53 promoter.
Nucleotide Number
DNA strand direction:5´
DNA strand direction: 3´
Nucleotide Number
DNA strand direction: 3´
DNA strand direction: 5´
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
In silico fingolimod and cladribine binding to p53 gene
Modified cladribine bind the p53 promoter via
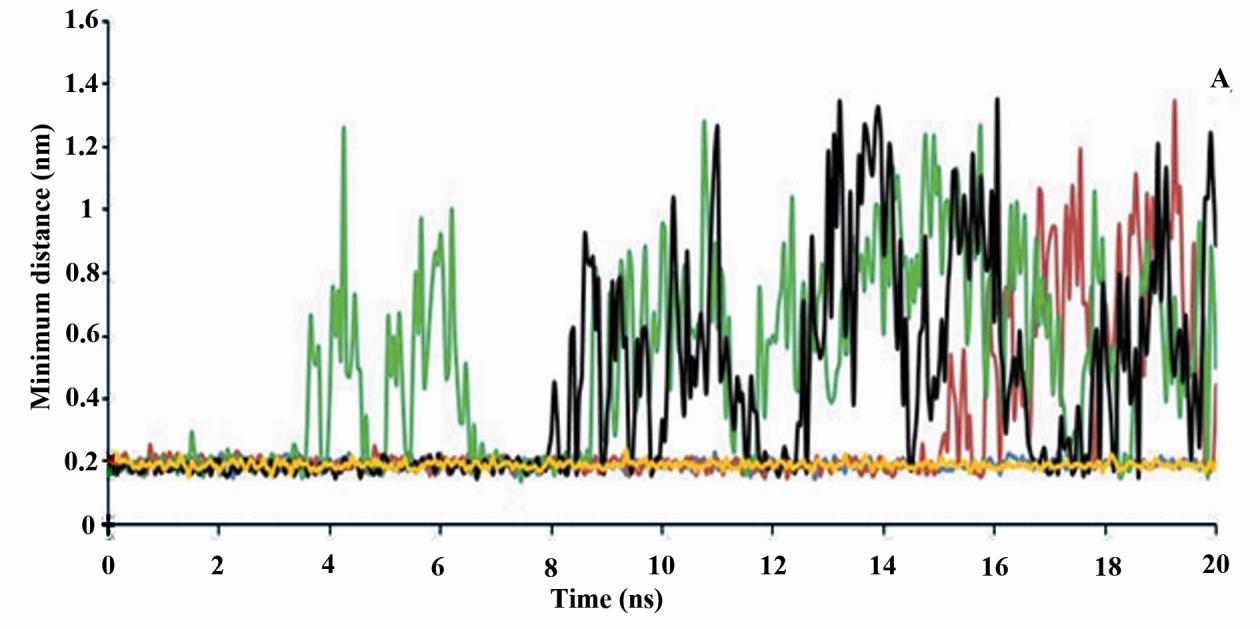
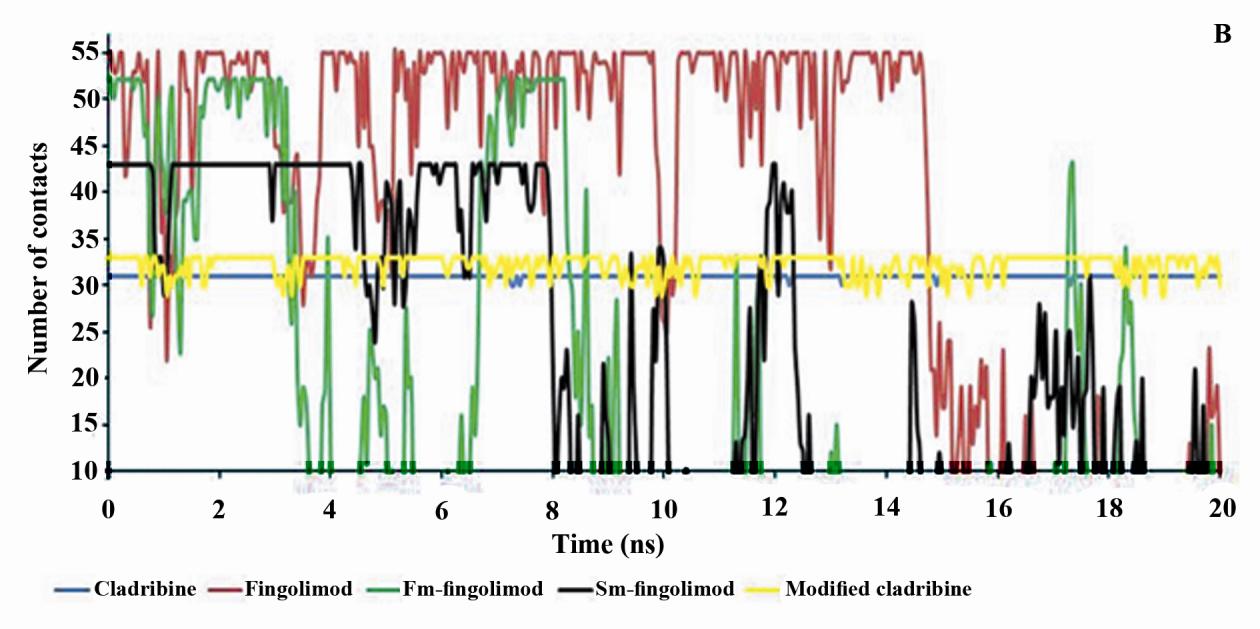
Minimum distance between p53 promoter and
its N2, N4, O1, C3, C7, C2, N3 and C8 atoms
ligands and the number of contacts less than 0.6
(Fig. 1A). In fingolimod and first modification
nm between p53 promoter and ligands during
only, three atoms (i.e. C16, C1 and C4) and in
the last five ns of MD simulations were also
second modified fingolimod only, three atoms
mentioned in Table 5. Figure 3 shows minimum
(i.e. C13, C5 and C8) (Fig. 1B) have SASA
distance between p53 promoter and ligands and
less than 10 Å2.
the number of contacts less than 0.6 nm
Table 5 shows the average number of hydrogen
between p53 promoter and ligands during the
bonds between ligands and the p53 promoter.
20 ns MD simulation.
Table 5. The average number of hydrogen bonds between ligands and p53 promoterand minimum distance
between them and number of contacts <0.6 nm between them during the last 5 ns of MD simulations
Average number of
Minimum distance
Number of contacts <0.6
hydrogen bonds between
between DNA and
nm between DNA and
DNA and drug
drug (nm)
Cladribine
Modified cladribie
Fingolimod
S m-fingolimod2
1. Fm-Fingolimod: First modification of fingolimod (i.e. deleting one carbon of fingolimod tail); 2. Sm-Fingolimod: Second modification of fingolimod (i.e. deleting four carbon of fingolimod tail).
The maximum number of hydrogen bonds
These results were confirmed by the minimum
p53 promoter belongs to
distance between ligands and p53 promoter
cladribine and modified cladribine, and this
and also the number of contacts between them
parameter is similar in them. Then their
(Fig. 3). In addition, first modified fingolimod
interactions with p53 promoter are strong
(fm-fingolomod) has the most minimum
(Table 5). In addition, the number of
distance and the least number of contacts with
hydrogen bonds between fingolimod, first or
p53 promoter among fingolimod and its
second modified fingolimod are the same but
derivatives. However, these parameters are
lower than those between cladribine and
more proper in second modification of
modified cladribine. This means that the
fingolimod (sm-fingolomod) and its interaction
interaction of fingolimod and its derivates
with p53 promoter is stronger relative to native
with p53 promoter is weak.
or first modified fingolimod (Table 5).
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
K. Mahnam et al.
Figure 3. The minimum distance (A) and the number of contacts less than 0.6 nm between p53 promoter and
drugs (B) during 20 ns of MD simulations. Fm-fingolimod: First modification of fingolimod,i.e. deleting one
carbon of fingolimod tail. Sm-fingolimod: Second modification of fingolimod, i.e. deleting four carbon of
fingolimod tail.
3. Binding free energy results
snapshots during the last 5 ns of MD
Table 6 shows binding free energy (ΔGb), Van
simulation. Binding free energy of cladribine,
der Waals and electrostatic energies of all
modified cladribine and second modified
ligands with p53 promoter obtained from 100
fingolimod to the p53 promoter is negative.
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
In silico fingolimod and cladribine binding to p53 gene
This means that these drugs can bind the p53
promoter and through inhibition of the p53
fingolimod and first modified fingolimod to
gene transcription probably induce cancer;
p53 promoter is positive, so they may not bind
then they can be supposedly carcinogen.
the p53 promoter.
Table 6. MM/PBSA binding free energies (kcal/mol) for ligand/DNA complexes during the last 5 ns of MD
simulation
Complex name
∆Eelec
∆Gpolar
∆Gbinding
Cladribine
Modified Cladribine
Fingolimod
S m-fingolimod2
1. Fm-Fingolimod: First modification of fingolimod (i.e. deleting one carbon of fingolimod t ail); 2. Sm-Fingolimod: Second modification of fingolimod (i.e. deleting four carbon of fingolimod tail). Abbreviations: ΔEelec = Electrostatic energy of interaction, ΔEvdw = Van der Waals energy of interaction. ∆Gpolar=polar
solvation free energy, ∆Gnon-polar= Non-polar solvation free energy.
Binding free energy of modified cladribine to
consistent with visual inspection of the middle
the p53 promoter is more positive and weaker
structures of the most populated structures
than native cladribine. The results obtained
obtained from MD simulation (Fig. 2).
from binding free energy (Table 6) and
MM/PBSA results show that binding of
docking (Table 1) for modified cladribine are
cladribine to the p53 promoter is more
opposite. Of course, results obtained from MD
negative than fingolimod which means that
simulation are more accurate than those from
cladribine probably is a powerful inhibitor in
dockings since water molecules and ions
initiation of p53 gene transcription. This may
explicitly present in molecular dynamics
be due to the similarity of purine rings of
simulation and MM/PBSA calculations, but in
cladribine to adenosine. The results of
dockings implicit solvent utilized and therefore
MM/PBSA calculations shows that as compare
water molecules and ions do not exist. This
with the native fingolimod, if one carbon is
suggests that MD simulation and MM/PBSA
(Fm-Fingolimod),
calculations are more accurate, and modified
binding free energy (ΔGb) increases but it
cladribine than to cladribine has a weaker
decreases when four carbons (sm-fingolimod)
interaction with p53 promoter.
are removed (Table 6). These results are
The negative binding free energy of the
consistent with MD simulation (Table 5 and
Fig. 3) but contrasted with docking results
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
K. Mahnam et al.
(Table1). Reducing four carbons from the
and more favorable for interactions of
aliphatic tails of fingolimod increases binding
fingolimod and its derivatives with p53
strength of fingolimod to the p53 promoter.
promoter (Table 6). This suggests that the
Then it is an inappropriate modification for
fingolimod and it can be investigated through
cladribine and fingolimod with p53 promoter
empirical studies. There is a very good
coordination between the average number of
The number of the first ten nucleotides with
hydrogen bonds during simulation and binding
the most total energy contributions in binding
free energy (Tables 5, 6). Also the differences
of ligands to the p53 promoter were mentioned
in the Van der Waals free and bound energies
in Table 7. As seen 3´-A89C88A90T87-5´ or
of all drugs during the last 5 ns MD simulation
5´-T16G17A18T19G20G21-3´ sequence has a
were calculated. According to the MM/PBSA
favorable interaction with cladribine however,
results, the Van der Waals interactions are
5´-G17A18T19G20G21-3´ sequence has a
more important (more negative) and more
favorable interaction with modified cladribine
favorable for interactions of cladribine and
(Tables 6 and 7). Interactions of fingolimod
modified cladribine with p53 promoter.
and its derivatives are weak and interaction
Electrostatic interactions are more important
energies are below -1.1 kcal/mol (Table 7).
Table 7.The first ten nucleotides that have the most total energy contribution in binding of drugs to p53
promoter (number of nucleotides are as mentioned in Table 4)
Cladribine
Modif ied cladribine
Fingolimod
Sm-f ingolimod2
Notes: 1.Fm-Fingolimod: First modification of fingolimod (i.e. deleting one carbon of fingolimod tail). 2. Sm-Fingolimod: Second modification of fingolimod (i.e. deleting four carbon of fingolimod tail). Num= Number of nucleotide in p53 promoter, Nuc=Nucleotide name, TE=Total energy of interaction each nucleotide with p53 promoter.
Conclusions
in the binding of cladribine and fingolimod and
In this in silico study we showed a difference
some of their derivatives to the p53 promoter.
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
In silico fingolimod and cladribine binding to p53 gene
This finding was confirmed by docking,
2-chloro-2´-deoxy
triphosphate (CldATP) (Foley et al., 2004) and
MM/PBSA methods.
fingolimod phosphate (Cohen et al., 2007) on
Based on the in silico studies it has been
p53 gene promoter since they are produced by
demonstrated that both cladribine and modified
some enzymes in the cell. Moreover, the effect
cladribine (replacing -OH on carbon 3´ ribose
of these drugs on exons of p53 gene is worth
sugar of adenosine with -CH3) can bind the
minor groove of p53 promoter and may lead to
conformational changes inp53 promoter. These
drugs can cause qualitative changes in the p53
The authors are grateful to Dr. Rashmi Kumari
for his help at installation of g_mmpbsa
carcinogenesis. MD simulation and MM/PBSA
module. The authors also wish to thank Miss
calculations showed that by modification of
Fateme Karimi for her assistance in preparing
cladribine its interactions decreases and the
modified cladribine may be less carcinogenic
than cladribine, assuming that the former
References
compound is a more favorable modification.
HJC, Postma JPM, Van
This phenomenon is explained by knowing the
Gunsteren WF, Dinola A, Haak JR. 1984.
increased cladribine size and steric prohibition
Molecular dynamics with coupling to an
with minor grove of p53 promoter. In addition,
external bath. J ChemPhys 81:3684-3690.
[2] BaiL, Zhu WG. 2006. p53: structure,
function and therapeutic applications. J
electrostatics interactions are more important
Cancer Mol 2(4):141-153.
for binding of cladribine to p53 promoter.
[3] Berendsen HJC, Van der Spoel D, Van
Removal of one carbon atom from the aliphatic
Drunen R. 1995. GROMACS: A message -
tails of fingolimod increased the binding free
energy whereas binding free energy decreased
by deletion of four carbon atoms. It is
Communications 91:43-56.
suggested that modifications in fingolimod or
[4] Baker NA, Sept D, Joseph S, Holst MJ,
cladribine structure may provide an interesting
McCammon JA. 2001. Electrostatics of
new direction for drug development. In the
nanosystems: Application to microtubules
future studies, it is suggested to investigate the
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
K. Mahnam et al.
98(18): 10037-10041.
Unrestrained 5 ns molecular dynamics
[5] Brown SP, Muchmore SW. 2009. Large-
simulation of a cisplatin-DNA 1, 2-GG
adduct provides a rationale for the NMR
Boltzmann surface area for routine physics-
conformational flexibility at the platinum
based scoring of protein-ligand complexes.
binding site. Journal of molecular biology
J Med Chem 52(10):3159-3165.
[6] Bradshaw RT, Patel BH, Tate EW,
[12] Eisenhaber F, Lijnzaad P, Argos P, Sander
Leatherbarrow RJ, Gould IR. 2011.
C, Scharf M. 1995. The Double Cube
Lattice Method: Efficient Approaches to
computational alanine scanning techniques
Numerical Integration of Surface Area and
forprobing a prototypical protein-protein
Volume and to Dot Surface Contouring of
interaction. Protein Eng Des Sel 24(1–2):
Molecular Assemblies J Comp Chem
B. 2007. Application of
Interferon Beta-1b in Multiple Sclerosis
American family physician 70:1935.
[8] Cheatham TE, Srinivasan J, Case DA,
[14] Foley TT, Hentosh P, Walters DE. 2004.
Kollman PA. 1998. Molecular dynamics
2-Chloro-2'-deoxyadenosine: alteration of
and continuum solvent studies of the
DNA: TATA element binding protein
stability of polyG-polyC and polyA-polyT
(TBP) interactions. Journal of molecular
DNA duplexes in solution. J Biomol Struct
modeling 10:32-37
Dyn 16(2):265-280.
[15] Frisch, M.J. et al., .1998. Gaussian 98
[9] Cohen B, Rieckmann P. 2007. Emerging
(Gaussian, Inc., Pittsburgh, PA).
oral therapies for multiple sclerosis.
[16] Gohlke H, Case DA. 2004. Converging
International journal of clinical practice
free energy estimates: MM-PB(GB)SA
studies on the protein-protein complex ras-
[10] Darden T, York D, Pedersen L. 1993.
raf. J ComputChem 25(2):238-250.
Particle meshEwald - An N.log (N)
[17] Hess B, Bekker H, Berendsen HJC, Fraaije
method for Ewald sums in large systems. J
JGEM. 1997. LINCS: a linear constraint
ChemPhys98:10089-10092.
solver for molecular simulations. J Comp
[11] Elizondo-Riojas MA, Kozelka J. 2001.
Chem18:1463-1472.
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
In silico fingolimod and cladribine binding to p53 gene
[18] Hess B, Kutzner D, Lindahl E. 2008.
[25] Lindahl E, Hess B, Van der Spoel D. 2001.
GROMACS 3.0: a package for molecular
efficient, load-balanced, and scalable
simulation and trajectory analysis. Journal
molecular simulation. J Chem Theory
of Molecular Modeling 7:306-317.
Comput 4:435-447.
[26] Morris G, Goodsell D, Halliday R, Huey R,
[19] Huo S, Massova I, Kollman PA. 2002.
Hart W, Belew R, Olson A. 1998.
Computational alanine scanning of the 1:1
Automated docking using a Lamarckian
human growth hormone-receptor complex.
genetic algorithm and an empirical binding
J ComputChem 23(1):15-27.
[20] Kar P, Lipowsky R, Knecht V. 2011.
computational chemistry 19(14):1639-1662.
Importance of polar solvation for cross-
reactivity of antibody and its variants with
Computational alanine scanning to probe
steroids. J PhysChem B 115:7661-7669.
[21] Kumari R, Kumar R, Consortium OSDD,
approach to evaluate binding free energies.
JACS 121(36):8133-8143.
GROMACS Tool for High-Throughput
[28] Majumdar R, Railkar R, Dighe RR. 2011.
Calculations. J ChemInf
Docking and free energy simulations to
Model Article ASAP.
predict conformational domains involved
[22] Karplus M, Kuriyan J. 2005. Molecular
in hCG-LH receptor interactions using
ProcNatlAcadSci 102(19):6679–6685.
Structure, Function, and Bioinformatics
79(11):3108-3122.
[29] Navikas V, Link H. 1996. Review:
YJ. 2011. Rotational viscosity calculation
cytokines and the pathogenesis of multiple
method for liquid crystal mixture using
sclerosis. Journal of neuroscience research
molecular dynamics 12(3):135-139.
[30] Neidle S, Pearl LH, Skelly JV. 1987. DNA
structure and perturbation by drug binding.
2008. Overexpression of USF increases
Biochemical Journal 243(1):1-13.
TGF-beta1 protein levels, but G1 phase
[31C, WFV. 2005.
arrest was not induced in FRTL-5 cells.
Free energies of ligand binding for
structurally diverse compounds. Proc Natl
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
K. Mahnam et al.
Acad Sci 102:6750-6754.
[32] Pronk S, Pãll S, Schulz R, Larsson P,
[33] Reisman D, Rotter V. 1993. The helix-loop-
Bjelkmar P, Apostolov R, et. al. 2013.
helix containing transcription factor USF
GROMACS 4.5: a high-throughput and
binds to and transactivates the promoter of
highly parallel open source molecular
the p53 tumor suppressor gene. Nucleic
simulation toolkit. Bioinformatics 29 (7):
acids research 21(2):345-350.
Molecular and Biochemical Diagnosis (MBD). Vol.1, No.2 (2014), 105-122
Source: http://mbd.modares.ac.ir/article_13300_810c3375e528cd566e23f73d608629f9.pdf
Injectable haloperidol is also available in an to confirmation bias, if it was mistakenly Pharmacies, Ambulatory Clinics, and Hospitals about alternative medications and any increases in Institute for Safe Medication Practices Canada A KEY PARTNER IN immediate-release formulation (haloperidol USP, as interpreted as meaning "lactate" (the salt present in
The Oncologist CME Program is located online at http://cme.theoncologist.com/. Symptom Management and Supportive Care The Assessment and Management of Delirium in Cancer Patients HIRLEY H. BUSH,a,b,c,d EDUARDO BRUERA aDepartment of Palliative Care & Rehabilitation Medicine, University of Texas M.D. Anderson Cancer






