Viagra gibt es mittlerweile nicht nur als Original, sondern auch in Form von Generika. Diese enthalten denselben Wirkstoff Sildenafil. Patienten suchen deshalb nach viagra generika schweiz, um ein günstigeres Präparat zu finden. Unterschiede bestehen oft nur in Verpackung und Preis.
Untitled
Journal of General Virology (2011), 92, 1172–1183
Reassortant low-pathogenic avian influenza H5N2viruses in African wild birds
Chantal J. Snoeck,13 Adeniyi T. Adeyanju,23 Se´bastien De Landtsheer,1Ulf Ottosson,2,3 Shiiwua Manu,2 Ward Hagemeijer,4 Taej Mundkur4and Claude P. Muller1
1Institute of Immunology, CRP-Sante´ and Laboratoire National de Sante´, Luxembourg
2A. P. Leventis Ornithological Research Institute, University of Jos, Jos, Plateau State, Nigeria
3Ottenby Bird Observatory, Degerhamn, Sweden
4Wetlands International, Wageningen, The Netherlands
To investigate the presence and persistence of avian influenza virus in African birds, we monitoredavian influenza in wild and domestic birds in two different regions in Nigeria. We found low-pathogenic avian influenza (LPAI) H5N2 viruses in three spur-winged geese (Plectropterusgambensis) in the Hadejia–Nguru wetlands. Phylogenetic analyses revealed that all of the genes,except the non-structural (NS) genes, of the LPAI H5N2 viruses were more closely related togenes recently found in wild and domestic birds in Europe. The NS genes formed a sister group toSouth African and Zambian NS genes. This suggested that the Nigerian LPAI H5N2 viruses foundin wild birds were reassortants exhibiting an NS gene that circulated for at least 7 years in Africanbirds and is part of the African influenza gene pool, and genes that were more recently introducedinto Africa from Eurasia, most probably by intercontinental migratory birds. Interestingly thehaemagglutinin and neuraminidase genes formed a sister branch to highly pathogenic avianinfluenza (HPAI) H5N2 strains found in the same wild bird species in the same wetland only1 year earlier. However, they were not the closest known relatives of each other, suggesting thattheir presence in the wetland resulted from two separate introductions. The presence of LPAIH5N2 in wild birds in the Hadejia–Nguru wetlands, where wild birds and poultry occasionally mix,
Received 13 December 2010
provides ample opportunity for infection across species boundaries, with the potential risk of
Accepted 18 January 2011
generating HPAI viruses after extensive circulation in poultry.
supports efficient short-range virus transmission of LPAIviruses by the faecal–oral route
Wild birds, in particular of the orders Charadriiformes and
High densities of mixed bird populations at
Anseriformes, are considered natural asymptomatic reser-
stopovers and non-breeding sites also promote intra- and
voirs of low pathogenic avian influenza (LPAI) virus
interspecies virus transmission ).
and the source ofinfluenza viruses in other species including poultry
Wetlands in Africa are preferred non-breeding sites for
All subtypes (H1–16 and N1–9) of
many Eurasian migratory waterbirds. Although Eurasian
avian influenza have been found in wild birds
species can mix with resident African birds or intra-African
Migration after the
migrants in many important bird areas ;
breeding season along distinct flyways contributes to long-
), only a few avian influenza viruses (AIV) have
distance dissemination of influenza viruses
been reported from Africa before 2006 (
and the aquatic environment of waterfowl
3These authors contributed equally to this paper.
The first seroprevalence study insub-Saharan Africa, conducted in commercial poultry in
The GenBank/EMBL/DDBJ accession numbers for the nucleotide
Nigeria between 1999 and 2004, did not detect antibodies to
sequences reported in this paper are FR771823–FR771846.
influenza viruses In a first wild-bird
Colour versions of Figs 1 and 2, showing phylogenies of the H5 and NS
surveillance effort, a wide variety of LPAI viruses were
genes, and six supplementary figures showing phylogenies of the NA,PA, PB1, PB2, matrix and NP genes are available with the online version
identified in migratory waterbirds
of this paper.
Interest in AIV in wild birds in Africa further increased in
029728 G 2011 SGM
Printed in Great Britain
LPAI H5N2 viruses in Nigerian wild birds
2006 when highly pathogenic avian influenza (HPAI) H5N1
the common teal (Anas crecca) from Egypt is not known
virus was first identified in Africa in Nigerian poultry farms
(After the independent introduction ofthree sublineages of clade 2.2 viruses into poultry, HPAI
In this study, we investigated the presence of avian
H5N1 viruses have undergone multiple reassortments and
influenza in wild and domestic birds in two different
reassorted viruses have largely replaced the initial sub-
locations in Nigeria in 2008. The Dagona wildlife sanctuary
lineages The route of introduction of
is a protected area within the Hadejia–Nguru wetlands,
H5N1 is unknown and both migratory birds and poultry
located in north-eastern Nigeria, and is part of the Chad
trade might be suspected
Basin national park. Large numbers of migratory birds mix
Although efforts to isolate influenza viruses
with local wild birds in the Hadejia–Nguru wetlands. The
from wild birds in Africa intensified, few met with success
Hadejia–Nguru wetlands are also the most important bird
area in the region, from which the first HPAI H5N1
probably been infected by scavenging H5N1-infected
outbreak in Africa was reported. The Amurum Forest
poultry but the infection route of
reserve is also a protected area located 15 km north-east of
Table 1. List of sampled domestic and wild bird species (order and family) in Dagona wildlife sanctuary and in the Amurum forestreserve and villages around Jos, Nigeria in 2008
For each family, the number of samples from domestic birds is given in parentheses.
Dagona wildlife sanctuary
Amurum Forest reserve and villages around Jos
No. species per family
No. samples per family
No. species per family
No. samples per family
C. J. Snoeck and others
Journal of General Virology 92
LPAI H5N2 viruses in Nigerian wild birds
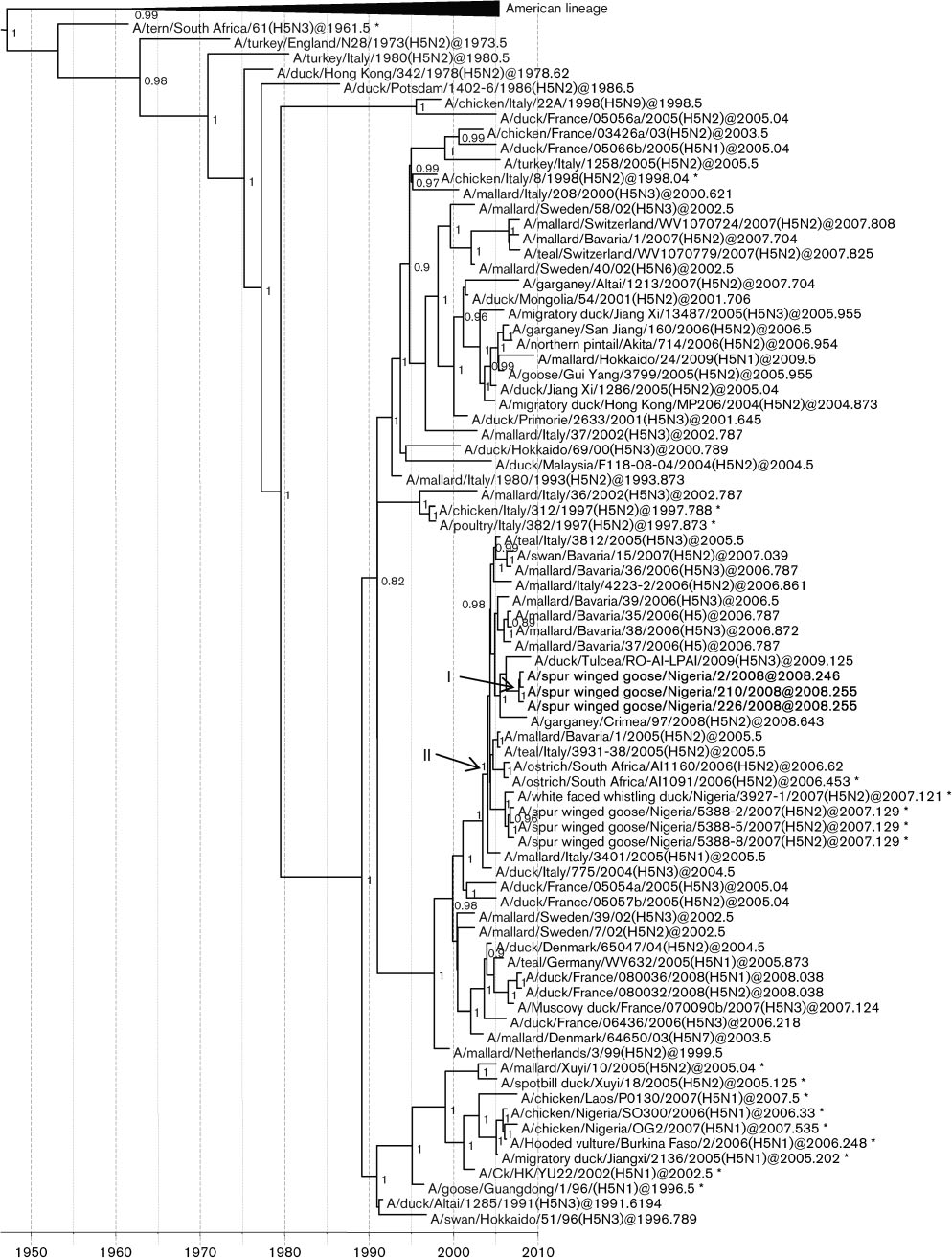
Fig. 1. Dated phylogeny of H5 genes. The horizontal axis represents calendar years and nodes correspond to mean TMRCAs.
Posterior probability values .0.75 are shown. Representative strains from Africa, Europe and Asia are included. Virus strainscharacterized in this study are shown in boldface type. *, HPAI strains. Node I represents the mean TMRCAs of the LPAI H5N2cluster. Node II corresponds to the most recent common ancestor of the Nigerian LPAI and HPAI supported by a high posteriorprobability value.
Jos (Plateau State), very close to the urban community
mammals [PB2 627E, 701D, 714S, PB1 678S, PA 615K and
). We did not find any HPAI H5N1 virus,
NP 319N (] or drug resistance [M2 26L,
but did find LPAI H5N2 strains that contained genes from
27V, 30A, 31S and 34G NA 119E (
the Eurasian gene pool and a non-structural (NS) gene that
was most closely related to other African viruses.
294N ] was found.
Interestingly the haemagglutinin (HA) genes were part ofa cluster that also contained HA genes from HPAI H5N2
Phylogenetic analyses of the eight genes reveal a
viruses found in the same bird species and in the same
separate origin for the NS gene
region 1 year earlier.
To explore the evolutionary origin and the time ofintroduction of LPAI H5N2 to Nigeria, tree topologies were
assessed and times to the most recent common ancestor(TMRCAs) were estimated using BEAST version 1.5.3. For
Avian influenza prevalence is low
each gene, the three LPAI H5N2 viruses were geneticallyhighly similar (Kimura distance from 0 to 0.9 %) and always
A total of 1024 samples were collected in the Dagona
clustered together (100 % posterior probabilities for all
wildlife sanctuary from 44 wild bird species, including
genes), suggesting that the three strains recently evolved
waterfowl, waders and passerines, and from 373 domestic
from a common ancestor and resulted from a single
poultry such as chickens, ducks and geese
introduction event in the Nigerian wetlands. All of the genes
Around Jos, cloacal samples were collected from 374
belonged to the Eurasian and not the American lineage, but
domestic birds (362 chickens, 5 ducks and 7 turkeys). In
none of them were related to HPAI H5N1 strains from
addition 154 wild birds corresponding to 52 species,
Nigeria, from other parts of Africa or from Eurasia.
mainly passerines, were sampled in the Amurum Forestreserve Only three fresh faecal samples collected
Phylogenetic analyses of the HA gene revealed that the
from spur-winged geese (Plectropterus gambensis) in a
LPAI H5N2 viruses were most closely related to a EuropeanH5N3 isolate (A/duck/Tulcea/RO-AI-LPAI/2009) recently
found in a sentinel duck in Romania The NA
(12u44.676 N, 10u40.001 E) were influenza A positive.
genes of the LPAI H5N2 viruses were most closely related
They were collected on 31 March 2008 (A/spur-winged
to an H2N2 strain from the Netherlands (A/mallard/
goose/Nigeria/2/2008) and on 3 April 2008 (A/spur-winged
Netherlands/14/2007) (Supplementary Fig. S1, available in
goose/Nigeria/210/2008 and A/spur-winged goose/Nigeria/
JGV Online). The LPAI H5N2 viruses formed a sister clade
226/2008). Thus, in our study, the prevalence of AIV in the
to strains recently isolated from wild birds in Europe (HA
Dagona wildlife sanctuary was 0.3 % (3/1024). All samples
and NA genes) and Asia (NA gene). Interestingly they also
collected in Plateau State were influenza A negative.
formed a sister branch with highly pathogenic H5N2strains that had been isolated one year earlier in a similar
Molecular analyses show no marker of virulence
location (10 km away) from the same wild bird species(HA and NA genes). In the same HA cluster, closely related
Genotype-specific PCRs were positive for H5 and N2 for the
HPAI and LPAI H5N2 viruses, both from South African
three viruses and all of the genes were sequenced using
ostriches (A/ostrich/South Africa/AI1160/2006, LPAI; and
previously published or newly designed primers. Analyses of
the HA gene showed that the predicted amino acid sequence
another sister branch to the Nigerian LPAI H5N2 viruses.
of the cleavage site of the three viruses corresponded to a low
However, all African isolates did not share a direct
pathogenic pathotype (PQRETR*GLF). They had a gluta-
common ancestor, suggesting that they did not directly
mine at position 226 and a glycine at position 228
evolve from each other. The TMRCAs of the LPAI H5N2
(numbering for the H3 subtype) indicating a higher binding
HA and NA genes ranged from February 2007 to March
affinity for sialic acid a2,3, which is characteristic of avian
2008 (95 % highest posterior density interval, HPD) and
cell-surface receptors (
October 2006 to March 2008 (95 % HPD), respectively.
No stalk deletion in the neuraminidase (NA) gene,nor additional predicted glycosylation sites, both proposed
The influenza virus PA polymerase subunit (PA) gene of
to be associated with poultry adaptation, were detected
the LPAI H5N2 viruses formed a sister clade with PA genes
from a goose and wild and captive ducks from France, the
genetic marker associated with increased virulence in
Netherlands and Sweden (Supplementary Fig. S2, available
C. J. Snoeck and others
Journal of General Virology 92
LPAI H5N2 viruses in Nigerian wild birds
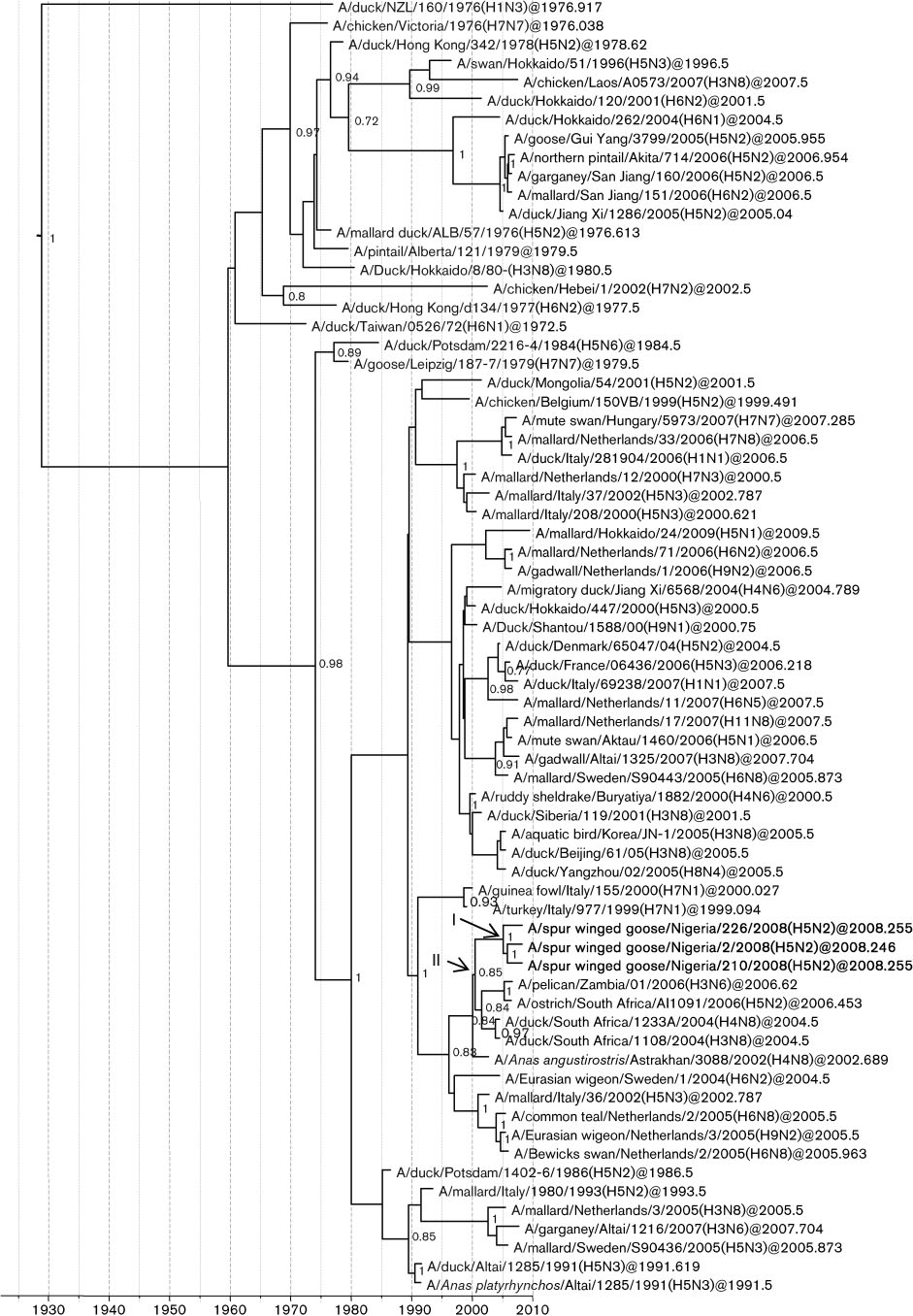
Fig. 2. Dated phylogeny of NS genes clustering in allele B. The horizontal axis represents calendar years and nodes correspondto mean TMRCAs. Posterior probability values .0.75 are shown. Representative strains from Africa, Europe and Asia areincluded. Virus strains characterized in this study are presented in boldface type. Node I represents the mean TMRCAs of theLPAI H5N2 cluster. Node II corresponds to the most recent common ancestor of the African NS genes, including the NigerianLPAI H5N2.
in JGV Online). The TMRCA of the LPAI H5N2 cluster
few exceptions (,
was estimated as being between July 2005 and February
2008 (95 % HPD). The polymerase basic protein 1 (PB1)
gene showed that the LPAI H5N2 viruses formed a sister
has received attention in Africa only after the introduction
clade with two wild-bird viruses from the Netherlands (A/
of HPAI H5N1 viruses. In our survey, three of 1024 (0.3 %)
samples collected in the Dagona wildlife sanctuary, a part
Netherlands/17/2007, H11N8) and a virus from one
of the Hadejia–Nguru wetlands, in north-eastern Nigeria
domestic goose from the Czech Republic (A/goose/Czech
were positive for influenza A. This corresponds to a
Republic/1848/2009, H7N9; Supplementary Fig. S3, avail-
prevalence of 1 % (3/312) in birds from the orders
able in JGV Online). The TMRCA of the LPAI H5N2
Charadriiformes and Anseriformes that are considered
cluster was estimated as being between June 2006 and
the natural reservoir of AIV In spur-
March 2008 (95 % HPD). The PB2 genes of the three
winged geese we found an infection rate of 2.4 % (3/123)
H5N2 strains were most closely related to A/quail/Italy/
compared with 8.2 % (8/97) in the earlier study
4610/2003 (H7N2) and the TMRCA of the LPAI H5N2
The three viruses found were low-pathogenic
cluster ranged from June 2005 to March 2008 (95 % HPD;
H5N2 viruses most closely related to each other over their
Supplementary Fig. S4, available in JGV Online).
full genomes. A tenfold higher prevalence (3.9 %) wasfound in waterbirds in the Hadejia–Nguru wetlands in the
The matrix gene sequences were more closely related to an
previous year This is similar to the
H9N2 matrix gene from the Netherlands (A/Bewick's
overall prevalence (3.5 %) found in a previous study in
swan/Netherlands/5/2007) and the TMRCA ranged from
wetlands throughout 12 African countries, including
June 2005 to January 2008 (95 % HPD; Supplementary Fig.
several neighbouring countries of Nigeria
S5, available in JGV Online). The three nucleoprotein (NP)
Many factors including the year, season, location,
genes clustered with viral genes isolated from four ducks,
species and age of birds influence the prevalence of AIV, as
one swan and one turnstone from Italy, the Netherlands,
is known from wild birds in Europe and America
Hungary, Germany and Sweden; the TMRCA of the cluster
formed by these three genes ranged from June 2005 to
February 2008 (95 % HPD; Supplementary Fig. S6,available in JGV Online).
Phylogenetic analyses revealed that all genes of our LPAIH5N2 viruses were most closely related to genes circulating
The NS gene sequences clustered in allele B and formed a
in the Eurasian wild-bird influenza gene pool. It has been
sister group to three South African NS sequences (A/
shown that migratory birds can carry LPAI viruses from
ostrich/South Africa/AI1091/2006, H5N2; A/duck/South
one continent to another ;
Africa/1108/2004, H3N8; and A/duck/South Africa/1233A/
introducing new genes and mixing gene pools
2004, H4N8) and to an H3N6 gene from Zambia (A/
pelican/Zambia/01/2006) Values larger than 95 %
Three main migratory flyways link Africa to Eurasia
HPD were observed for the TMRCA of the Nigerian LPAI
(the East Atlantic flyway, the Black Sea–Mediterranean
H5N2 cluster (January 2002 to February 2007), which may
flyway and the East Africa–West Asia flyway;
indicate an older origin for that gene, although we cannot
and Nigeria is located where they intersect. Thus, low
exclude that our dataset for the NS segment does not
pathogenic avian influenza genes may have been intro-
contain sufficient phylogenetic signal to provide precise
duced from Eurasia to Africa by migratory birds. However,
estimates. Also, the exclusion of the distantly related NS
spur-winged geese are in principle a sedentary sub-Saharan
gene of A/duck/NZL/160/1976 (H1N3) from the phylo-
species, normally making only short daily flights, and only
genetic analysis had little influence on the TMRCA. The
rarely longer flights, depending on the availability of water.
close relationship suggests that all seven viruses shared a
The species is widespread throughout sub-Saharan Africa,
recent common ancestor that was introduced to Africa
but does not leave the continent
(TMRCA of the African NS cluster, node II in
During the dry season, spur-winged geese
November 1997 to May 2003).
are highly gregarious around permanent waterbodiessuch as in the Hadejia–Nguruwetlands where they mix with Afro-tropical birds and
Eurasian migratory birds such as garganey (Anas querque-
Although avian influenza has been extensively studied for
dula), northern pintail (Anas acuta) or ferruginous duck
decades in wild birds from the northern hemisphere, with
(Aythya nyroca) (
C. J. Snoeck and others
The mingling of Eurasian migratory and
from the Eurasian lineages have been introduced into
African bird species in the Hadejia–Nguru wetlands seems
North America and gradually replaced the American H6
to be reflected also in the AIV gene pools.
Interestingly, the NS gene sequences of the three LPAI
The presence of LPAI H5N2 viruses in African wild birds
H5N2 viruses from Nigeria shared a common ancestor
represents a certain risk also for poultry. Infected wild
with South African and Zambian influenza isolates from
birds, such as spur-winged geese, may introduce LPAI
2004 and 2006, suggesting another origin for the NS gene,
viruses into free-ranging domestic ducks reared in the
in contrast to all of the other genes. Also, the TMRCA of
wetlands in north-eastern Nigeria with whom they
the Nigerian LPAI H5N2 NS genes suggested that the
occasionally mix ). Similar situations
cluster emerged in approximately October 2004 (January
have already been observed in South Africa where wild
2002–February 2007; 95 % HPD; node I in whereas
birds were suspected to have infected domestic birds with
the mean TMRCAs for the other genes were found to be
whom they shared similar genes Farmed ostriches
during late 2006 or 2007 The TMRCA of the
were infected with H6N8 in 1998 and
African monophyletic cluster, which included the Nigerian,
2007 with H5N2 viruses in 2004 and
South African and Zambian strains, suggested that a virus
was introduced to Africa between 1997 and 2003. This
H9N2 in 2008. H10N7 was also found in domestic ducks
ancestral virus evolved and probably reassorted with other
) In addition, HPAI viruses
viruses present in the African wild bird population, as has
are thought to emerge after extensive circulation of H5
already been suggested
(and H7) LPAI subtypes and adaptation in poultry
The NS gene was acquired
by viruses that later spread throughout Africa. Ring
This is of particular concern since
recoveries have shown that some intra-African migratory
the Nigerian LPAI H5N2 strains belong to a genetic cluster
birds from South Africa [e.g. comb duck (Sarkidiornis
that seems to have an increased propensity to develop the
melanotos)] sometimes migrate as far north as West Africa
highly pathogenic phenotype. Indeed, from a common
and thus would be able to transmit
node (node II, that emerged between June 2002 and
AIV over long distances within Africa. The presence of
May 2004 (95 % HPD), highly pathogenic H5N2 strains
similar genes over a four year period (2004–2008;
emerged twice in South Africa in 2004 (not shown in
and its probable introduction at least 7 years ago suggests
because of its shorter sequence; and in
that AIV can persist in the African bird population.
2006 and in Nigeria in 2007
The Eurasian–African AIV interface resembles the situationin the Bering Strait where interregional transmission of
Interestingly, the latter Nigerian HPAI H5N2 viruses were
influenza viruses occurs between North American and
found only 1 year earlier, about 10 km away in the same
Eurasian birds. Genes from the Asian lineage have been
wetlands and in the same wild bird species as the LPAI
found in Alaska )
H5N2 described in this study. For both the HA and NA
and genes from the American lineages have been found in
genes, the HPAI H5N2 formed a sister branch to the LPAI
Japan producing a variety of reassortants.
H5N2 but were not the closest known relatives of each
Also, similarly to the persistence of Eurasian genes in
other, suggesting that their presence in the wetlands
Africa, now constituting the African gene pool, H6 genes
resulted from two separate introductions. The question
Table 2. Estimated TMRCAs of the Nigerian LPAI H5N2 cluster, nucleotide substitution model and sequence length used in theBayesian analyses
General time-reversible (GTR) substitution model with a gamma (C) and invariant (I) site heterogeneity model.
Mean TMRCA (95 % HPD)
Nucleotide substitution model
Sequence length (nt)
March 2007 (Jun 05 – Mar 08)
June 2007 (Jun 06 – Mar 08)
March 2007 (Jul 05 – Feb 08)
October 2007 (Feb 07 – Mar 08)
January 2007 (Jun 05 – Feb 08)
GTR+I+C with simplifications: CP3.cg5CP3.gt,
August 2007 (Oct 06 – Mar 08)
December 2006 (Jun 05 – Jan 08)
GTR+I+C with simplifications: CP3.cg5CP3.at,
October 2004 (Jan 02 – Feb 07)
GTR+C with simplifications: CP3.cg5CP3.gt
Journal of General Virology 92
LPAI H5N2 viruses in Nigerian wild birds
Fig. 3. Genetic relationship of African AIV viruses, other than HPAI H5N1 viruses, sequenced since 2004, by year, in wild anddomestic birds. Genes have the same colour code if they share a direct common ancestor. Unsequenced genes are indicatedas dotted bars and genes that have no African sister gene are shown as white bars. Phylogenetic relationships were firstassessed by comparing the African strains with all avian influenza strains downloaded on to the NCBI Influenza Virus Resourcedatabase, but based on shorter fragments, depending on the shortest sequences available for the African strains.
Representative strains were selected for each gene based on these preliminary analyses and trees were calculated using MEGA4by using the neighbour-joining method and by using the Kimura two parameters model and 1000bootstrap replicates. *, Values corresponding to the probability values from The figure suggests, based on the availabledata (from South Africa, Nigeria and Zambia), that some avian influenza viruses may be maintained in the African wild-birdpopulation where reassortment events can occur, and that these viruses can be transmitted from wild to domestic birds. Only A/DK/SA/811/04 (LPAI H5N1) does not share any gene with a domestic bird strain. OS, Ostrich; EG, Egyptian goose; SWG,spur-winged goose; WFWD, white-faced whistling duck; PD, pekin duck; PE, pelican; SA, South Africa.
C. J. Snoeck and others
also remains as to where the Nigerian HPAI H5N2 virus
were predicted for HA and NA by using the NetNGlyc 1.0 server
acquired its HPAI phenotype. The wetlands in north-
(http://www.cbs.dtu.dk/services/NetNGlyc/). Kimura distances were
eastern Nigeria provide ample opportunity for cross-
calculated with MEGA4 (using the Kimura twoparameters model. For each gene, phylogenetic relationships were
species infection and perhaps even the generation of
inferred by comparing the LPAI H5N2 strains with all avian influenza
HPAI viruses, which normally only occur after circulation
strains downloaded to the NCBI Influenza Virus Resource database
and adaptation in poultry.
(up until 15 December 2009; www.ncbi.nlm.nih.gov./genomes/FLU/)after removing short sequences and sequences with insertions or
In conclusion, we report the presence of LPAI H5N2
deletions resulting in frame shifts. Datasets were aligned using
viruses in wild birds in an African wetland, which were
CLUSTAL W Coding regions were used for
reassortants with genes from the Eurasian and African gene
phylogenetic analyses and only the first ORFs were used for the
pools, as strong evidence of the introduction of low-
matrix and NS genes. Trees were calculated using MEGA4 (
pathogenic avian influenza into Africa by Eurasian
with the neighbour-joining method by using the Kimura
migratory birds. Furthermore, the circulation of LPAI
two parameters model and 1000 bootstrap replicates. Representativestrains were selected for each gene based on these preliminary
and HPAI H5N2 strains in wild birds in African wetlands
that emerged from a cluster that had an increasedpropensity to develop the highly pathogenic phenotype
Tree topologies, substitution rates and TMRCAs were estimated by aBayesian Markov-chain Monte Carlo (MCMC) method
represents a high risk for poultry, especially in areas with
implemented in BEAST version 1.5.3
low biosecurity that provide opportunities for cross-species
Depending on the available details on isolation
dates, the exact isolation dates, the mid-month dates (15th), the mid-interval dates or the mid-year dates were used as calibration points.
For each dataset, different substitution models with two codon
partitions to allow independent estimates for the third codonposition, two uncorrelated relaxed-clock models (log-normal and
Wild bird surveillance. Wild birds were captured with mist nets in
exponential distributions; and two
the Dagona wildlife sanctuary in north-eastern Nigeria between 28
coalescent models (constant population size and Bayesian skyline;
March and 22 April 2008. Oropharyngeal and cloacal swabs as well as
were compared visually in TRACER version
fresh faecal samples were collected after ensuring the species of origin.
1.5.3 and statistically using a Bayes
Domestic poultry in the villages around the wetlands were also
factor test (the ratio of the marginal likelihoods of two models)
sampled. All samples were collected in triplicate with cotton swabs,
as implemented in TRACER
stored in virus transport medium [PBS pH 7.0 with 2000 U
version 1.5.3, in order to identify the model that fitted the data best.
streptomycin ml-1,
Evidence against the null model, which is the model with the lowest
B ml-1, 250 mg gentamicin ml-1, 60 mg ofloxacin ml-1, 200 mg
marginal likelihood, was assessed by the method proposed by
sulfamethoxazole ml-1 and 2.5 mg amphotericin B ml-1] and placed
When there was no evidence against the null model or
directly into liquid nitrogen in the field. In addition, between 4
when this evidence was weak, the simplest model was kept to avoid
December 2007 and 5 March 2008, swabs were collected from wild
unnecessary overparameterization. Analyses revealed that the GTR
birds in the Amurum forest reserve (Plateau State) and from backyard
substitution model, assuming an uncorrelated exponential relaxed
poultry in five villages around Jos.
clock and a constant population size, was the model that best fittedthe data for all genes. For matrix, NP and NS genes, the GTR model
RNA Extraction, RT-PCR and sequencing. RNA was extracted
was further simplified to avoid overparameterization Two
from 50 ml of virus transport medium by using a MagMAX-96 AI/ND
to three runs of 50–1006106 generations of the MCMC method were
Viral RNA Isolation kit (Ambion) and a KingFisher 96 (Thermo
performed and sampled to produce 10 000 trees each. Convergence of
Fisher). Influenza A-positive specimens were detected by using a real-
the runs was confirmed in TRACER version 1.5.3. The results of
time RT-PCR assay targeting the matrix gene and previously
multiple runs were combined using LogCombiner version 1.5.3
published gene-specific primers and probe
with a burn-in of 10–25 %,
RT-PCRs were carried out using the following
summarized into the maximum clade credibility tree using
cycling conditions: reverse transcription for 30 min at 50 uC,
TreeAnnotator version 1.5.3 and
denaturation at 95 uC for 15 min followed by 40 cycles of
visualized in FigTree version 1.3.1
amplification at 95 uC for 10 s, 60 uC for 20 s. Amplifications wereperformed with a Qiagen OneStep RT-PCR kit using 2 ml of RNA in afinal volume of 25 ml. Matrix-positive samples were tested for H5
H7 (http://www.defra.gov.uk/vla/science/docs/sci_ai_vi536.pdf) and N1 genotypes. The
The authors wish to thank A. Sausy and E. Charpentier for technical
eight genes were then amplified by using several PCRs targeting
help, D. Kihlberg, P. O
¨ sterman, A. Eriksson, Dr Ullarama and the
overlapping fragments (primer sequences and details available upon
A. P. Leventis Ornithological Research Institute (APLORI) for their
request). PCR products were purified using a JetQuick PCR
expertise in sample collection and the Nigerian Authority for
Purification Spin kit (Genomed). Sequencing was performed as
authorization and support. They also acknowledge the Wild Bird
previously described using PCR primers as
Global Avian Influenza Network for Surveillance (GAINS) project of
sequencing primers.
the Global Health Program, Wildlife Conservation Society, funded byUSAID and Wetlands International, for financial support. They
Molecular and phylogenetic analyses. Sequence assembly and
gratefully acknowledge the Ministry of Cooperation of Luxembourg,
analyses were performed using SeqScape version 2.5 (Applied
the Ministry of Health, the Ministry of Research and the Centre de
Biosystems) and BioEdit The nucleotide sequences are
Recherche Public-Sante´ for their generous financial and moral
available in the GenBank/EMBL/DDBJ databases under the accession
support. C. J. S. was supported by an AFR fellowship from the
numbers FR771823–FR771846. Potential N-linked glycosylation sites
Fonds National de la Recherche, Luxembourg. This study was also
Journal of General Virology 92
LPAI H5N2 viruses in Nigerian wild birds
supported by the contribution No. 242 from Ottenby Bird
Drummond, A. J., Ho, S. Y., Phillips, M. J. & Rambaut, A. (2006).
Observatory and contribution No. 43 from APLORI.
Relaxed phylogenetics and dating with confidence. PLoS Biol 4, e88.
Ducatez, M. F., Olinger, C. M., Owoade, A. A., De Landtsheer, S.,Ammerlaan, W., Niesters, H. G., Osterhaus, A. D., Fouchier, R. A. &
Muller, C. P. (2006). Avian flu: multiple introductions of H5N1 inNigeria. Nature 442, 37.
Abolnik, C. (2007a). Molecular characterization of H5N2 avian
Ducatez, M. F., Olinger, C. M., Owoade, A. A., Tarnagda, Z., Tahita,
influenza viruses isolated from South African ostriches in 2006. Avian
M. C., Sow, A., De Landtsheer, S., Ammerlaan, W., Ouedraogo, J. B. &
Dis 51, 873–879.
other authors (2007a). Molecular and antigenic evolution and
Abolnik, C. (2007b). Molecular Epidemiology of Newcastle Disease and
geographical spread of H5N1 highly pathogenic avian influenza viruses
Avian Influenza in South Africa, pp. 286. Pretoria: University of
in western Africa. J Gen Virol 88, 2297–2306.
Pretoria Department of Zoology and Entomology.
Ducatez, M. F., Tarnagda, Z., Tahita, M. C., Sow, A., de Landtsheer, S.,
Abolnik, C., Cornelius, E., Bisschop, S. P., Romito, M. & Verwoerd, D.
Londt, B. Z., Brown, I. H., Osterhaus, D. M., Fouchier, R. A. & other
(2006). Phylogenetic analyses of genes from South African LPAI
authors (2007b). Genetic characterization of HPAI (H5N1) viruses from
viruses isolated in 2004 from wild aquatic birds suggests introduction
poultry and wild vultures, Burkina Faso. Emerg Infect Dis 13, 611–613.
by Eurasian migrants. Dev Biol (Basel) 124, 189–199.
Dugan, V. G., Chen, R., Spiro, D. J., Sengamalay, N., Zaborsky, J.,
Abolnik, C., Bisschop, S. P., Gerdes, G. H., Olivier, A. J. & Horner,
Ghedin, E., Nolting, J., Swayne, D. E., Runstadler, J. A. & other
R. F. (2007). Phylogenetic analysis of low-pathogenicity avian
authors (2008). The evolutionary genetics and emergence of avian
influenza H6N2 viruses from chicken outbreaks (2001-2005) suggest
influenza viruses in wild birds. PLoS Pathog 4, e1000076.
that they are reassortants of historic ostrich low-pathogenicity avian
Ezealor, A. U. (2001). Nigeria. In Important Bird Areas in Africa and
influenza H9N2 and H6N8 viruses. Avian Dis 51 (Suppl.), 279–284.
Associated Islands: Priority Sites for Conservation, pp. 673–692. Edited
Abolnik, C., Londt, B. Z., Manvell, R. J., Shell, W., Banks, J., Gerdes,
by L. D. C. Fishpool & M. I. Evans. Newbury and Cambridge: Pisces
G. H., Akol, G. & Brown, I. H. (2009). Characterisation of a highly
Publications & BirdLife International.
pathogenic influenza A virus of subtype H5N2 isolated from ostriches
Fouchier, R. A., Munster, V., Wallensten, A., Bestebroer, T. M.,
in South Africa in 2004. Influenza Other Respir Viruses 3, 63–68.
Herfst, S., Smith, D., Rimmelzwaan, G. F., Olsen, B. & Osterhaus, A. D.
Abolnik, C., Gerdes, G. H., Sinclair, M., Ganzevoort, B. W., Kitching,
(2005). Characterization of a novel influenza A virus hemagglutinin
J. P., Burger, C. E., Romito, M., Dreyer, M., Swanepoel, S. & other
subtype (H16) obtained from black-headed gulls. J Virol 79, 2814–
authors (2010). Phylogenetic analysis of influenza A viruses (H6N8,
H1N8, H4N2, H9N2, H10N7) isolated from wild birds, ducks, and
Fouchier, R. A. M., Munster, V. J., Keawcharoen, J., Osterhaus,
ostriches in South Africa from 2007 to 2009. Avian Dis 54 (Suppl.),
A. D. M. E. & Kuiken, T. (2007). Virology of avian influenza in relation
to wild birds. J Wildl Dis 43, S7–S14.
Alexander, D. J. (2007). An overview of the epidemiology of avian
Fusaro, A., Joannis, T., Monne, I., Salviato, A., Yakubu, B., Meseko, C.,
influenza. Vaccine 25, 5637–5644.
Oladokun, T., Fassina, S., Capua, I. & Cattoli, G. (2009). Introduction
Alexander, D. J. & Brown, I. H. (2000). Recent zoonoses caused by
into Nigeria of a distinct genotype of avian influenza virus (H5N1).
influenza A viruses. Rev Sci Tech 19, 197–225.
Emerg Infect Dis 15, 445–447.
Gabriel, G., Dauber, B., Wolff, T., Planz, O., Klenk, H. D. & Stech, J.
Allwright, D. M., Burger, W. P., Geyer, A. & Terblanche, A. W. (1993).
Isolation of an influenza A virus from ostriches (Struthio camelus).
(2005). The viral polymerase mediates adaptation of an avian
Avian Pathol 22, 59–65.
influenza virus to a mammalian host. Proc Natl Acad Sci U S A102, 18590–18595.
Baigent, S. J. & McCauley, J. W. (2003). Influenza type A in humans,
Gaidet, N., Dodman, T., Caron, A., Balanc¸a, G., Desvaux, S.,
mammals and birds: determinants of virus virulence, host-range and
Goutard, F., Cattoli, G., Lamarque, F., Hagemeijer, W. & Monicat, F.
interspecies transmission. Bioessays 25, 657–671.
(2007). Avian influenza viruses in water birds, Africa. Emerg Infect Dis
Banks, J., Speidel, E. C., McCauley, J. W. & Alexander, D. J. (2000).
13, 626–629.
Phylogenetic analysis of H7 haemagglutinin subtype influenza A
Gaidet, N., Cattoli, G., Hammoumi, S., Newman, S. H., Hagemeijer, W.,
viruses. Arch Virol 145, 1047–1058.
Takekawa, J. Y., Cappelle, J., Dodman, T., Joannis, T. & other authors
Becker, W. B. (1966). The isolation and classification of Tern virus:
(2008). Evidence of infection by H5N2 highly pathogenic avian
influenza virus A/Tern/South Africa/1961. J Hyg (Lond) 64, 309–320.
influenza viruses in healthy wild waterfowl. PLoS Pathog 4, e1000127.
Brown, L. H., Urban, E. K. & Newman, K. (1982). The Birds of Africa,
Garamszegi, L. Z. & Møller, A. P. (2007). Prevalence of avian
vol. I. London: Academic Press.
influenza and host ecology. Proc Biol Sci 274, 2003–2012.
Gubareva, L. V., Kaiser, L., Matrosovich, M. N., Soo-Hoo, Y. &
Slingenbergh, J. (2008). Agro-ecological features of the introduction
Hayden, F. G. (2001). Selection of influenza virus mutants in
and spread of the highly pathogenic avian influenza (HPAI) H5N1 in
experimentally infected volunteers treated with oseltamivir. J Infect
northern Nigeria. Geospat Health 3, 7–16.
Dis 183, 523–531.
Drummond, A. J. & Rambaut, A. (2007). BEAST: Bayesian evolutionary
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence
analysis by sampling trees. BMC Evol Biol 7, 214.
alignment editor and analysis program for Windows 95/98/NT.
Drummond, A. J., Nicholls, G. K., Rodrigo, A. G. & Solomon, W.
Nucleic Acids Symp Ser 41, 95–98.
(2002). Estimating mutation parameters, population history and
Hay, A. J., Collins, P. J. & Russell, R. J. (2008). Antivirals and
genealogy simultaneously from temporally spaced sequence data.
resistance. In Avian influenza, pp. 252–271. Edited by H.-D. Klenk,
Genetics 161, 1307–1320.
M. N. Matrosovich & J. Stech. Basel: Karger.
Drummond, A. J., Rambaut, A., Shapiro, B. & Pybus, O. G. (2005).
Hockey, P. A. R., Dean, W. R. J. & Ryan, P. G. (2005). Roberts - Birds of
Bayesian coalescent inference of past population dynamics from
Southern Africa, 7th edn. Cape Town: The Trustees of the John
molecular sequences. Mol Biol Evol 22, 1185–1192.
Voelcker Bird Book Fund.
C. J. Snoeck and others
Ip, H. S., Flint, P. L., Franson, J. C., Dusek, R. J., Derksen, D. V., Gill, R.
Pfitzer, S., Verwoerd, D. J., Gerdes, G. H., Labuschagne, A. E.,
E. J., Jr, Ely, C. R., Pearce, J. M., Lanctot, R. B. & other authors (2008).
Erasmus, A., Manvell, R. J. & Grund, C. (2000). Newcastle disease and
Prevalence of Influenza A viruses in wild migratory birds in Alaska:
avian influenza A virus in wild waterfowl in South Africa. Avian Dis
patterns of variation in detection at a crossroads of intercontinental
44, 655–660.
flyways. Virol J 5, 71.
ProMED (2006). Avian influenza - worldwide: Nigeria, OIE. Accessed
Ito, T., Okazaki, K., Kawaoka, Y., Takada, A., Webster, R. G. & Kida, H.
9 April 2009 at http://www.promedmail.org/, archive number
(1995). Perpetuation of influenza A viruses in Alaskan waterfowl
reservoirs. Arch Virol 140, 1163–1172.
Rambaut, A. (2009). FigTree v1.3.1. Accessed 10 March 2010 at http://
Kass, R. E. & Raftery, A. E. (1995). Bayes Factors. J Am Stat Assoc 90,
Rambaut, A. & Drummond, A. J. (2009). Tracer v1.5. Accessed 20
Kiso, M., Mitamura, K., Sakai-Tagawa, Y., Shiraishi, K., Kawakami, C.,
March 2010 at http://tree.bio.ed.ac.uk/software/tracer/.
Kimura, K., Hayden, F. G., Sugaya, N. & Kawaoka, Y. (2004). Resistant
Ramey, A. M., Pearce, J. M., Flint, P. L., Ip, H. S., Derksen, D. V.,
influenza A viruses in children treated with oseltamivir: descriptive
Franson, J. C., Petrula, M. J., Scotton, B. D., Sowl, K. M. & Wege, M. L.
study. Lancet 364, 759–765.
(2010). Intercontinental reassortment and genomic variation of low
Koehler, A. V., Pearce, J. M., Flint, P. L., Franson, J. C. & Ip, H. S.
pathogenic avian influenza viruses isolated from northern pintails
(2008). Genetic evidence of intercontinental movement of avian
(Anas acuta) in Alaska: examining the evidence through space and
influenza in a migratory bird: the northern pintail (Anas acuta). Mol
time. Virology 401, 179–189.
Ecol 17, 4754–4762.
Rogers, G. N., Paulson, J. C., Daniels, R. S., Skehel, J. J., Wilson, I. A.
Le, Q. M., Kiso, M., Someya, K., Sakai, Y. T., Nguyen, T. H., Nguyen,
& Wiley, D. C. (1983). Single amino acid substitutions in influenza
K. H., Pham, N. D., Ngyen, H. H., Yamada, S. & other authors (2005).
haemagglutinin change receptor binding specificity. Nature 304, 76–
Avian flu: isolation of drug-resistant H5N1 virus. Nature 437, 1108.
Liu, J. H., Okazaki, K., Bai, G. R., Shi, W. M., Mweene, A. & Kida, H.
¨ hm, C., Horimoto, T., Kawaoka, Y., Su¨ss, J. & Webster, R. G. (1995).
(2004). Interregional transmission of the internal protein genes of H2
Do hemagglutinin genes of highly pathogenic avian influenza viruses
influenza virus in migratory ducks from North America to Eurasia.
constitute unique phylogenetic lineages? Virology 209, 664–670.
Virus Genes 29, 81–86.
Saad, M. D., Ahmed, L. S., Gamal-Eldein, M. A., Fouda, M. K., Khalil, F.,
Lupiani, B. & Reddy, S. M. (2009). The history of avian influenza.
Yingst, S. L., Parker, M. A. & Montevillel, M. R. (2007). Possible avian
Comp Immunol Microbiol Infect Dis 32, 311–323.
influenza (H5N1) from migratory bird, Egypt. Emerg Infect Dis 13,1120–1121.
Matrosovich, M. N., Gambaryan, A. S., Teneberg, S., Piskarev, V. E.,Yamnikova, S. S., Lvov, D. K., Robertson, J. S. & Karlsson, K. A.
Scott, D. A. & Rose, P. M. (1996). Atlas of Anatidae Populations in
(1997). Avian influenza A viruses differ from human viruses by
Africa and Western Eurasia. Wageningen: Wetlands International.
recognition of sialyloligosaccharides and gangliosides and by a higher
Simulundu, E., Mweene, A. S., Tomabechi, D., Hang'ombe, B. M.,
conservation of the HA receptor-binding site. Virology 233, 224–234.
Ishii, A., Suzuki, Y., Nakamura, I., Sawa, H., Sugimoto, C. & other
Matrosovich, M., Zhou, N., Kawaoka, Y. & Webster, R. (1999). The
authors (2009). Characterization of H3N6 avian influenza virus
surface glycoproteins of H5 influenza viruses isolated from humans,
isolated from a wild white pelican in Zambia. Arch Virol 154, 1517–
chickens, and wild aquatic birds have distinguishable properties.
J Virol 73, 1146–1155.
Snoeck, C. J., Ducatez, M. F., Owoade, A. A., Faleke, O. O., Alkali,
Munster, V. J. & Fouchier, R. A. (2009). Avian influenza virus: of virus
B. R., Tahita, M. C., Tarnagda, Z., Ouedraogo, J. B., Maikano, I. &
and bird ecology. Vaccine 27, 6340–6344.
other authors (2009). Newcastle disease virus in West Africa: newvirulent strains identified in non-commercial farms. Arch Virol 154,
Munster, V. J., Wallensten, A., Baas, C., Rimmelzwaan, G. F.,
Schutten, M., Olsen, B., Osterhaus, A. D. & Fouchier, R. A. (2005).
Mallards and highly pathogenic avian influenza ancestral viruses,
Spackman, E., Stallknecht, D. E., Slemons, R. D., Winker, K., Suarez,
northern Europe. Emerg Infect Dis 11, 1545–1551.
D. L., Scott, M. & Swayne, D. E. (2005). Phylogenetic analyses of typeA influenza genes in natural reservoir species in North America
Munster, V. J., Baas, C., Lexmond, P., Waldenstro
¨ m, J., Wallensten, A.,
reveals genetic variation. Virus Res 114, 89–100.
Fransson, T., Rimmelzwaan, G. F., Beyer, W. E., Schutten, M. & otherauthors (2007). Spatial, temporal, and species variation in prevalence
Stallknecht, D. E., Shane, S. M., Zwank, P. J., Senne, D. A. & Kearney,
of influenza A viruses in wild migratory birds. PLoS Pathog 3, e61.
M. T. (1990). Avian influenza viruses from migratory and residentducks of coastal Louisiana. Avian Dis 34, 398–405.
Olsen, B., Munster, V. J., Wallensten, A., Waldenstro¨m, J., Osterhaus,A. D. & Fouchier, R. A. (2006). Global patterns of influenza a virus in
Suarez, D. L. (2000). Evolution of avian influenza viruses. Vet
wild birds. Science 312, 384–388.
Microbiol 74, 15–27.
Owoade, A. A., Ducatez, M. F. & Muller, C. P. (2006). Seroprevalence of
Suchard, M. A., Weiss, R. E. & Sinsheimer, J. S. (2001). Bayesian
avian influenza virus, infectious bronchitis virus, reovirus, avian
selection of continuous-time Markov chain evolutionary models. Mol
pneumovirus, infectious laryngotracheitis virus, and avian leukosis virus
Biol Evol 18, 1001–1013.
in Nigerian poultry. Avian Dis 50, 222–227.
Tamura, K., Dudley, J., Nei, M. & Kumar, S. (2007). MEGA4: molecular
Owoade, A. A., Gerloff, N. A., Ducatez, M. F., Taiwo, J. O., Kremer, J. R.
evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol
& Muller, C. P. (2008). Replacement of sublineages of avian influenza
Evol 24, 1596–1599.
(H5N1) by reassortments, sub-Saharan Africa. Emerg Infect Dis 14,
Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). CLUSTAL W:
improving the sensitivity of progressive multiple sequence alignment
through sequence weighting, position-specific gap penalties and
wantanapokin, S., Buranathai, C., Amonsin, A., Theamboonlers, A.
weight matrix choice. Nucleic Acids Res 22, 4673–4680.
& Poovorawan, Y. (2006). Single step multiplex real-time RT-PCR for
Wallensten, A., Munster, V. J., Latorre-Margalef, N., Brytting, M.,
H5N1 influenza A virus detection. J Virol Methods 131, 143–147.
Elmberg, J., Fouchier, R. A., Fransson, T., Haemig, P. D., Karlsson, M.
Journal of General Virology 92
LPAI H5N2 viruses in Nigerian wild birds
& other authors (2007). Surveillance of influenza A virus in migratory
Webster, R. G., Bean, W. J., Gorman, O. T., Chambers, T. M. &
waterfowl in northern Europe. Emerg Infect Dis 13, 404–411.
Kawaoka, Y. (1992). Evolution and ecology of influenza A viruses.
Ward, C. L., Dempsey, M. H., Ring, C. J., Kempson, R. E., Zhang, L.,
Microbiol Rev 56, 152–179.
Gor, D., Snowden, B. W. & Tisdale, M. (2004). Design and
zu Dohna, H., Li, J., Cardona, C. J., Miller, J. & Carpenter, T. E. (2009).
performance testing of quantitative real time PCR assays for influenza
Invasions by Eurasian avian influenza virus H6 genes and replacement
A and B viral load measurement. J Clin Virol 29, 179–188.
of the virus' North American clade. Emerg Infect Dis 15, 1040–1045.
Source: http://www.access.ottenby.se/meddelanden/reports/242.pdf
JIDA_FebMarch2008 30/01/2008 16:12 Page 1 Volume 54 Number 1 February/March 2008 Journal of the Irish Dental AssociationIris Cumainn Déadach na hÉireann Oral care in juvenile JIDA_FebMarch2008 30/01/2008 16:14 Page 29 Journal of the Irish Dental Association Oral health and orthodonticconsiderations in children with juvenileidiopathic arthritis: review of theliterature and report of a case
Semantic Data Platform for Healthcare Lead beneficiary: MUG D3.1 Sketch of system Date: 31/03/2014 architecture specification Nature: Report WP3 – Architecture and Dissemination level: PU D3.1 – Sketch of system architecture specification WP3: Architecture and Requirements Dissemination level: Public Authors: Philipp Daumke, Carla Haid, Luke Mertens (Averbis),





