Viagra gibt es mittlerweile nicht nur als Original, sondern auch in Form von Generika. Diese enthalten denselben Wirkstoff Sildenafil. Patienten suchen deshalb nach viagra generika schweiz, um ein günstigeres Präparat zu finden. Unterschiede bestehen oft nur in Verpackung und Preis.
Npgrj_nbt_1481 1.8
A small molecule enhances RNA interference andpromotes microRNA processing
Ge Shan1,6, Yujing Li1,6, Junliang Zhang2, Wendi Li1, Keith E Szulwach1, Ranhui Duan1,Mohammad A Faghihi3, Ahmad M Khalil3, Lianghua Lu2, Zain Paroo4, Anthony W S Chan1,Zhangjie Shi5, Qinghua Liu4, Claes Wahlestedt3, Chuan He2 & Peng Jin1
Small interfering RNAs (siRNAs) and microRNAs (miRNAs) are sequence-specific post-transcriptional regulators of geneexpression. Although major components of the RNA interference (RNAi) pathway have been identified, regulatory mechanismsfor this pathway remain largely unknown. Here we demonstrate that the RNAi pathway can be modulated intracellularly by smallmolecules. We have developed a cell-based assay to monitor the activity of the RNAi pathway and find that the small-molecule
enoxacin (Penetrex) enhances siRNA-mediated mRNA degradation and promotes the biogenesis of endogenous miRNAs. Weshow that this RNAi-enhancing activity depends on the trans-activation-responsive region RNA-binding protein. Our results
provide a proof-of-principle demonstration that small molecules can be used to modulate the activity of the RNAi pathway. RNAienhancers may be useful in the development of research tools and therapeutics.
RNAi is a well-conserved mechanism that uses small noncod-
ing RNAs to silence gene expression post-transcriptional
A chemical screen to identify small molecules that enhance RNAi
Gene regulation by RNAi has been recognized as one of the major
To alter the activity and gain insight into the regulation of the RNAi
regulatory pathways in eukaryotic cellThe endogenous small
pathway, we developed a reporter system to monitor RNAi activity.
RNAs can shape diverse cellular pathways, including chromo-
In this system, a stable cell line derived from human embryonic
some architecture, development, growth control, apoptosis and stem
kidney (HEK293) cells, expressing a gene encoding 293-EGFP
lishing Gr
(enhanced green fluorescent protein), was infected with a lentivirus
RNAi operates via two post-transcriptional mechanisms: targeted
expressing a short hairpin RNA (shRNA) that is processed into siRNA
mRNA degradation by siRNA and suppression of translation/degrada-
specifically targeting EGFP mRNA (Fig. 1a)The transduction led to
tion by miRNA. The RNAi mechanism has been co-opted by
reduced levels of EGFP in 293-EGFP cells; these cells are called RNAi-
researchers and has achieved broad utility in gene-function analysis,
293-EGFP in the experiments following. To verify that the siRNAs
drug-target discovery and validation, and therapeutic developmen
against EGFP reduced EGFP expression, we transfected the clones with
2008 Nature Pub
Given the pivotal roles of endogenous small RNAs in diverse biological
2-O-methyl-RNAs, which has been shown to block the activity of the
pathways and the broad application of RNAi in biology and
lentivirus-encoded EGFP siRNAand we observed increased GFP
medicine, understanding the mechanism of the RNAi pathway is of
expression (Fig. 1a). To reduce variation between experiments, we
great importance.
isolated individual cell clones with moderate reductions in GFP
Over the last several years, key protein components involved
expression and used them to screen for both inhibitors and enhancers
in the RNAi pathway have been identified; however, little is known
of the RNAi pathway.
about the regulation of the RNAi pathway itself. Here we describe a
Using this system, we screened a collection of 2,000 US Food and
chemical biology approach to modulate the RNAi pathway and report
Drug Administration–approved compounds and natural products
the identification of a small molecule that enhances RNAi and
and identified a small molecule named enoxacin that enhanced
promotes the biogenesis of miRNA by facilitating the interaction
siRNA-mediated mRNA degradation. This small molecule was
between trans-activation-responsive region RNA-binding protein
enoxacin (Fig. 1a). Enoxacin increased siGFP-mediated gene
(TRBP) and RNAs. Our results provide a proof-of-principle demon-
knockdown mediated by siRNA against EGFP in our cell-based
stration that small molecules can be used to understand what cellular
reporter system in a dose-dependent manner, with a median effective
factors affect the activity of the RNAi pathway.
concentration (EC50) of B30 mM, whereas it had no effect on the
1Department of Human Genetics, Emory University School of Medicine, 615 Michael St., Atlanta, Georgia 30322, USA. 2Department of Chemistry, The University ofChicago, 929 East 57th St., Chicago, Illinois 60637, USA. 3Department of Molecular and Integrative Neurosciences, The Scripps Research Institute, 5353 Parkside Drive,Jupiter, Florida 33458, USA. 4Department of Biochemistry, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, Texas 75390, USA.
5The College of Chemistry, Peking University, 202 Chengfu Rd., Beijing, 100871, P.R. China. 6These authors contributed equally to this work. Correspondence should beaddressed to P.J. (
Received 8 January; accepted 24 June; published online 20 July 2008;
NATURE BIOTECHNOLOGY
ADVANCE ONLINE PUBLICATION

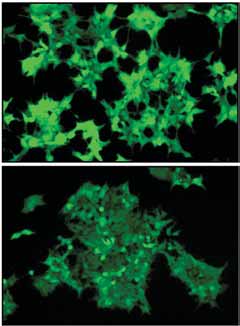
293-EGFP RNAi-293-EGFP
Enoxacin (50 µM)
+Enoxacin (50 µM)
Library of 2,000 small molecules
Relative quantification
of EGFP protein (%)
Enhancer of RNAi (enoxacin)
Enoxacin (50 µM)
Figure 1 Identification of a small molecule enhancing RNAi through a
chemical screen. (a) HEK293 cells stably expressing EGFP (293-EGFP)
were infected with lentivirus producing shRNA against EGFP (shRNA-
EGFP); the resulting RNAi-293-EGFP cells with reduced GFP expression
mRNA knockdown (%)
were isolated. The RNAi-293-EGFP cells transfected with 2-O-methyl
Relative GAPDH mRNA
RNA against the GFP siRNA are shown in the middle with the recovery
10–13 10–12 10–11 10–10 10–9 10–8 10–7
of GFP expression. A mutant 2-O-methyl RNA against the GFP siRNA is
BACE1 siRNA concentration (M)
shown on the right as a negative control. The RNAi-293-EGFP cells were
used for a chemical screen, which led to the identification of an RNAi
shVector + enoxacin
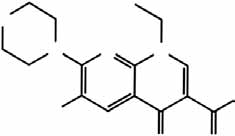
enhancer (enoxacin). The chemical structure of enoxacin is shown
shGAPDH + enoxacin
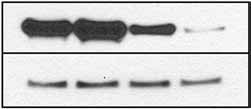
on the right. (b) Enoxacin enhances shRNA-EGFP-mediated genesilencing. The GFP protein levels were detected by western blot analysis using anti-EGFP antibody (right), with GAPDH as a loading control. The
quantification is shown below. Fluorescence images of RNAi-293-EGFP cells without (top) or with (bottom) enoxacin are shown on the left (exposuretime is different from panel a). Values are mean ± s.d. (c) Enoxacin enhances shGAPDH-mediated gene silencing. Relative GAPDH mRNA levelsin cells are determined by quantitative RT-PCR. Values are mean ± s.d. for triplicate samples. *, P o 0.001. (d) Enoxacin potentiates syntheticsiRNA-induced knockdown of BACE1. Synthetic siRNA against human BACE1 was transfected at a range of concentrations, from 1 pM to 20 nM, inHEK293FT cells. Knockdown of BACE1 mRNA was graphed as a percentage of mock-treated samples in the presence or absence of enoxacin. Values are
mean ± s.d. for triplicate samples.
cells expressing GFP only (Fig. 1b and Supplementary Fig. 1).
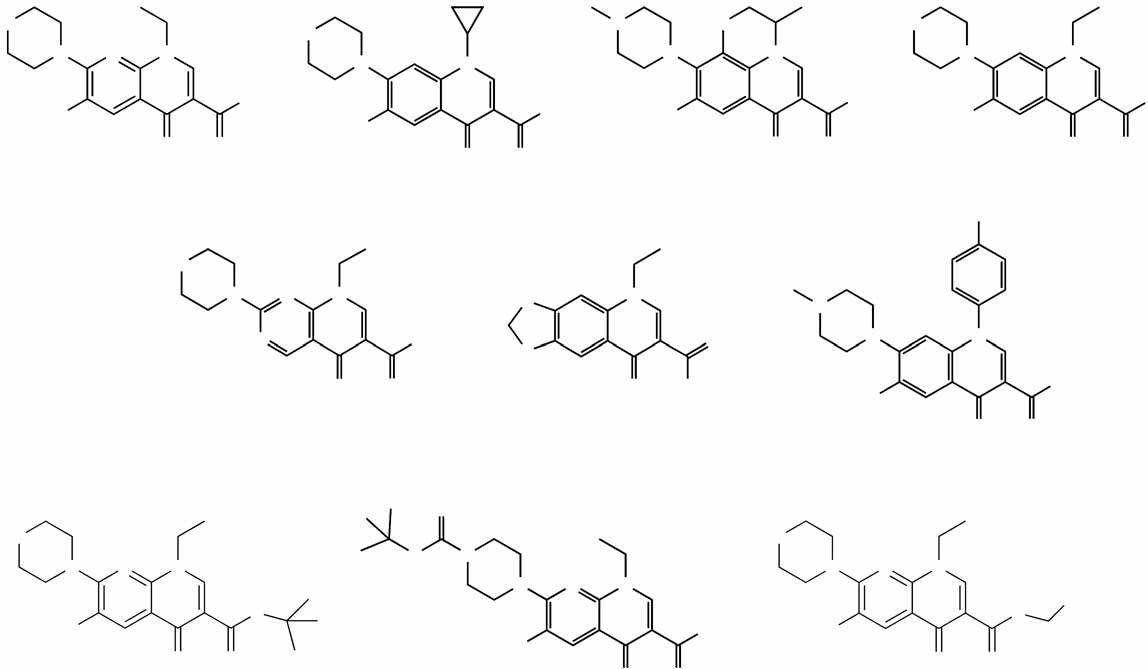
RNAi-enhancing activity of enoxacin is structure dependent
Importantly, enoxacin was relatively nontoxic, even at the high
Enoxacin belongs to a family of synthetic antibacterial compounds
lishing Gr
concentration of 150 mM, which is lower than a clinical doseSimilar
based on a fluoroquinolone skelet. Fluoroquinolones have a broad
enhancement of gene knockdown mediated by GAPDH-specific
antimicrobial spectrum and are very successful at treating a variety of
shRNA (shGAPDH) in cells stably expressing an shGAPDH was also
bacterial infection. As a family, fluoroquinolones target bacterial
observed upon enoxacin treatment (Fig. 1c). We also tested other
type II topoisomerases, such as DNA gyrase in Gram-negative bacteria
shRNAs against GFP (different from the GFP used in our reporter
and DNA topoisomerase IV in Gram-positive bacter; these agents
system), luciferase and Fmr1, and again observed comparable
do not inhibit eukaryotic topoisomerase II. Enoxacin has been used to
increases (Supplementary Fig. 2), indicating that the effect of enox-
treat bacterial infections ranging from gonorrhea to urinary tract
2008 Nature Pub
acin on RNAi is indeed universal.
infectionClinically, side effects have been minimal in adult
There are currently two means of harnessing the RNAi machinery
To test whether the quinolone family in general acts to enhance the
to induce specific suppression of gene expression in cells: shRNAs
RNAi pathway, we examined the effects of several other quinolones,
and siRNA duplexes. Hence we also questioned whether enox-
both commercially available and synthetically modified molecules
acin would have any effect on siRNA duplex–induced RNA
(enoxacin-V1-3), using our RNAi GFP reporter system (Fig. 3a).
We found that enoxacin consistently gave rise to a left-shift of
We found that two of these compounds (ciprofloxacin (Cipro) and
the concentration-response curve to an siRNA specifically target-
norfloxacin (Noroxin)) have substantial RNAi-enhancing activity;
ing human BACE1 mRNA (Fig. 1d). Importantly, the levels of
however, these two molecules are less effective than enoxacin itself
unrelated transcripts, such as those encoding PINK-1 and actin,
(Fig. 3b). Most other commercially available quinolones tested had
were not changed, suggesting that enoxacin did not induce
either much less or almost no RNAi-enhancing activity. The addition
nonspecific effects (data not shown). We also observed similar
of sterically blocking groups to the C-3 carboxylate (enoxacin-V1 and
enhancement of knockdown efficiency by enoxacin using siRNA
enoxacin-V3) and the piperazine terminal nitrogen atom of enoxacin
duplexes against different genes in various cell lines (Fig. 2).
(enoxacin-V2) reduced the RNAi-enhancing activity. Substitutions at
Furthermore, by comparing the gene knockdown efficiency among
the N-1, C-6 and C-7 positions of enoxacin also interfered with its
different concentrations of siRNA duplex used for transfection, we
RNAi-enhancing activity. The sensitivity of the RNAi-enhancing
found that enoxacin substantially reduced the siRNA dosage required
activity of enoxacin to chemical substitution suggests that it forms a
to achieve comparable knockdown efficiency (Figs. 1d and 2). These
specific complex distinct from the known targets of quinolones.
data together suggest that the small-molecule enoxacin enhances RNAi
To exclude the possibility that enoxacin enhances RNAi activity by
induced by either shRNAs or siRNA duplexes and substan-
increasing the expression of one or more components in the RNAi
tially reduces the dosage required to achieve gene knockdown in
pathway, we first examined the protein levels of multiple components
mammalian cells.
in the RNA-induced silencing complex (RISC) and found no
ADVANCE ONLINE PUBLICATION
NATURE BIOTECHNOLOGY
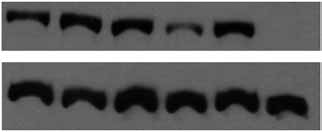
siRNA against mouse GAPDH b
siRNA against human Htt
siRNA against Her2
siRNA against Aha1
siRNA against PP1A
Control 10 nM 5 nM
siRNA against MAPK1
siRNA against EGFP
siRNA against KIF11
siRNA against Actin
siRNA against human
Figure 2 Enoxacin potentiates additional synthetic siRNA-induced gene knockdown. (a) NIH3T3 cells were transfected with different amounts of siRNAduplexes against mouse GAPDH in the absence or presence of enoxacin. Relative GAPDH mRNA levels in the cells transfected with control and differentdosages of siRNA are determined by quantitative RT-PCR. (b) 293 cells stably expressing Htt were transfected with 10 or 20 nM siRNA duplexes against
human Htt in the absence or presence of enoxacin. The amounts of Htt are determined by western blot. (c) MCF-7 cells were transfected with 5 nM siRNAduplexes against human Her2 in the absence or presence of enoxacin. Relative Her2 mRNA levels are determined by TaqMan assay. (d) 293 cells weretransfected with 5 nM siRNA duplexes against human Aha1 in the absence or presence of enoxacin. Relative Aha1 mRNA levels are determined by TaqMan
assay. (e) 293 cells were transfected with 5 nM siRNA duplexes against human PP1A in the absence or presence of enoxacin. Relative PP1A mRNA levelsare determined by TaqMan assay. (f) 293 cells were transfected with 5 nM siRNA duplexes against human MAPK1 in the absence or presence of enoxacin.
Relative MAPK1 mRNA levels are determined by TaqMan assay. (g) 293-EGFP cells were transfected with 5 nM siRNA duplexes against EGFP in theabsence or presence of enoxacin. Relative EGFP mRNA levels are determined by quantitative RT-PCR. (h) 293 cells were transfected with 5 nM siRNA
duplexes against human KIF11 in the absence or presence of enoxacin. Relative KIF11 mRNA levels are determined by TaqMan assay. (i) HeLa cells weretransfected with 20 nM siRNA duplexes against human actin in the absence or presence of enoxacin. Relative actin mRNA levels are determined by TaqMan
assay. (j) 293 cells were transfected with 5 nM siRNA duplexes against human GAPDH in the absence or presence of enoxacin. Relative GAPDH mRNAlevels are determined by TaqMan assay. In panels a and c–j, values are mean ± s.d.
lishing Gr
alteration in these levels in the cells treated with enoxacin (Supple-
enoxacin on global gene expressionWe performed microarray
mentary Fig. 3). Because previous studies have shown that fluoro-
analyses using both HEK293 and mouse NIH3T3 cells. Only a small
quinolones could massively increase or reduce steady-state levels of
number of genes (36 out of B22,000 genes expressed in HEK293 cells
multiple mRNAs at a much higher concentration (250 mM) than we
and 10 out of B20,000 genes expressed in NIH3T3 cells) displayed
used in this study (50 mM), we further investigated the effect of
significant changes (41.6-fold, P o 0.001) in enoxacin-treated
2008 Nature Pub
Oxolinic acid (OA)
Relative RNAi-enhancing activity (%)
Oxolinic acid (OA)
Figure 3 Determination of chemical structure required for RNAi-enhancing activity. (a) The chemical structures of enoxacin variants are shown. (b) RelativeRNAi-enhancing activities of enoxacin variants are shown. RNAi-enhancing activity was determined using RNAi-293-EGFP cells and fluorescencequantification on an Analyst HT plate reader. After subtraction of background fluorescence, the reduction of GFP fluorescence by enoxacin was set as 100%.
Relative fluorescence intensity reductions measured in cells treated with the other compounds were normalized to the fluorescence reduction by enoxacin.
Values are mean ± s.d.
NATURE BIOTECHNOLOGY
ADVANCE ONLINE PUBLICATION
Figure 4 Enoxacin promotes the processing of miRNAs and the loading of
siRNA duplexes onto RISCs. (a) Northern blots show that enoxacin enhances
the level of siRNAs and mature miR-125a in cells transfected with shRNA
against GFP, luciferase and miR-125a plasmids, respectively. 5S RNA was
used as loading control. (b) Quantitative RT-PCR was used to measure the
levels of pri-, pre- and mature forms of miR-125a, miR-124a and miR-199b
in mock- and enoxacin-treated (50 mM) HEK293 cells. Values are mean ±
s.d. for triplicate samples. (c) miRNAs with consistent changes in enoxacin-treated HEK293 cells (transfected HEK293 cells stably expressing pri-miR-
125a were used). Each miRNA was examined by TaqMan MiRNA assay, andmock-treated cells are used as the baseline for all comparisons. The bar
graph shows the average fold change with s.d. of the miRNAs that displayed
consistent changes in triplicate experiments. (d) Top panel shows the
western blot to detect the myc-Ago2 fusion protein in both input and
immunoprecipitated complex (IP) using anti-myc antibody. Northern blots
detecting the guide strand and passenger strand of siRNA duplexes are
shown in the middle. (Note that different probes were used to detect the
guide and passenger strands of siRNA duplexes, so the signal intensities
appearing on the blot do not reflect the absolute amount of each strand.)
The intensity of each band is quantified and indicated under the blot
92 ± 9 100 250 ± 20 100
(untreated Input and untreated IP are 100). Bottom panel shows the relative
amount of small RNAs associated with the immunoprecipitated Ago2-
105 ± 8 100 154 ± 12 100
containing RISCs in the presence or absence of enoxacin (75 mM), which
was normalized to the inputs. Endogenous miR-30a that was not affected by
Relative expression level (%)
enoxacin was used as a control (miR-30e shown in panel c is transcribed
from a different genomic locus from miR-30a). Values are
mean ± s.d.
associated with Ago2 (%)
of endogenous miRNAs (142/157) were not significantly affected.
Relative amount of small RNAs
Pri+Pre-miR Mature-miR
Most of the miRNAs altered by enoxacin (13/15) had approximately
twofold increases in expression of the mature form, whereas only two
of the miRNAs had decreased levels of the mature forms (Fig. 4c). Aswith miR-125a, we also found decreased levels of the primary and
cells, and we were unable to find any gene consistently in both cell
precursor forms of the miRNAs whose mature forms increased in the
types (Supplementary Tables 1 and 2). These results suggest that
presence of enoxacin (Fig. 4b and Supplementary Fig. 5). Interest-
lishing Gr
the RNAi-enhancing activity we observed with enoxacin is not
ingly, we noted that the precursor forms of the elevated endogenous
caused by pleiotropic effects on gene expression. We therefore
miRNAs are generally abundant in untreated HEK293 cells, whereas
hypothesized that enoxacin must interact specifically with the nucleic
the endogenous miRNAs that are not significantly affected by enox-
acids and protein(s) involved in the RNAi pathway to increase RNAi-
acin generally have very little or undetectable precursor in cells
enhancing activity.
(Supplementary Table 3). Furthermore, enoxacin treatment couldincrease the production of mature small RNAs from the miR-30a–
Enoxacin promotes processing of miRNAs and loading of siRNAs
based shRNAs (pre-miRNA-like RNAs) that were abundant in cells,
2008 Nature Pub
To determine the biological target(s) of enoxacin and the mechanism
whereas enoxacin had no effect on endogenous miR-30a, which had
by which enoxacin modulates the RNAi pathway, we examined the
no detectable steady-state precursor form in cells (Fig. 4a; siLuciferase
expression of mature small RNAs in cells stably expressing either
and data not shown). These data suggest that enoxacin could promote
different shRNAs or the primary transcript of miR-125a (pri-miR-
the processing of pre-miRNAs. This enhancement, mediated by
125a). We observed consistent increases in the expression of small
enoxacin, depends largely on the amount of precursor RNAs in
RNAs in cells treated with enoxacin, despite the use of different
cells, rather than specific RNA sequences.
promoters (Fig. 4a and Supplementary Fig. 4). Furthermore, we
We also determined the effect of enoxacin on the stability and
found that the addition of enoxacin led to increases in the mature
loading of siRNA duplexes onto RISC. It has been shown that
form of miR-125a and corresponding decreases in the level of pri-
Argonaute2 (Ago2) is a key component of the RISC responsible
miR-125a alone, as well as decreases in the total level of pri-miR-125a
for mRNA cleavage activity (Slicer activity. We isolated Ago2-
and precursor miR-125a (pre-miR-125a) (Fig. 4a–b and Supplemen-
containing RISCs through immunoprecipitation from HEK293 cells
tary Fig. 5). This suggests that enoxacin can promote the processing of
transfected with siRNA duplexes and determined the amounts of
miR-125a. The shGFP used in our initial reporter system mimics pre-
siRNAs associated with Ago2 in the presence or absence of enoxacin
miRNAs; therefore, it is processed by Dicer, rather than Drosha, which
using quantitative RT-PCR. The addition of enoxacin had no effect on
processes pri-miRNAs to pre-miRNAsFurthermore, enoxacin
the amount of siRNAs in the input, suggesting that enoxacin does not
exerted effects on the siRNA duplex, which likewise does not require
simply enhance the stability of siRNAs in vivo (Fig. 4d). With a similar
DroshaThese data suggest that enoxacin might function at the level
amount of Ago2 protein immunoprecipitated, the amount of trans-
of Dicer-mediated precursor processing and/or loading onto RISCs.
fected siRNAs associated with Ago2-containing RISCs increased two-
To examine the effect of enoxacin on endogenous miRNAs, we used
fold upon treatment with enoxacin, whereas endogenous miR-30a,
miRNA TaqMan assays to monitor the profiles of 157 miRNAs in
which is not altered by enoxacin, showed no difference (Fig. 4d, top
transfected HEK293 cells stably expressing pri-miR-125a. The majority
and bottom panels).
ADVANCE ONLINE PUBLICATION
NATURE BIOTECHNOLOGY
Enoxacin K :94 nM
Relative cleavage
Relative amount of
Percentage of RNAs bound by
Control Enoxacin Control Enoxacin
Time of binding reactions (minutes)
Relative RNAi-enhancing activity
Relative GAPDH mRNA level
Figure 5 Enoxacin facilitates the TRBP-RNA interaction, and the RNAi-enhancing
activity is TRBP-dependent. (a) Left panel shows in vitro processing of
Relative GAPDH mRNA
radioactively labeled let-7 precursor by either Dicer alone or Dicer with TRBP.
Arrows indicate the precursor and processed miRNAs. Right panel shows the
averages of relative cleavage activity from 4 independent experiments (N ¼ 4).
*, P o 0.001 when the reaction with Dicer, TRBP and enoxacin was compared
with other reactions. Values are mean ± s.d. (b) RNA-binding assay with
recombinant TRBP protein and 5¢-32P-labeled let-7 precursor in the presence
or absence of enoxacin (30 mM) is shown in the top panel (RNA-binding time:
Relative RNAi-enhancing
60 min). Bottom left panel shows the percentage of let-7 precursor bound by
Level of TRBP mRNA:
recombinant TRBP over time in the presence or absence of RNAi-E (30 mM).
*, P o 0.001. Bottom right panel shows the percentage of let-7 precursor boundby recombinant TRBP versus log of the protein concentration in the presence orabsence of enoxacin (30 mM). Values are mean ± s.d. (c) The RNAi-enhancing activity is TRBP dependent. The siRNAs against TRBP were transfected intothe cells stably expressing shGAPDH used in Figure 1c in the presence or absence of enoxacin (50 mM). The reduction of TRBP had no effect on shGAPDH-
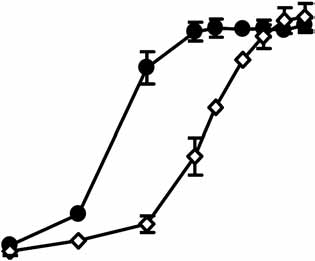
mediated mRNA degradation because the GAPDH mRNA level remained unchanged in the absence of enoxacin. The correlation between TRBP mRNA leveland RNAi-enhancing activity is shown. Values are mean ± s.d.
Because quantitative RT-PCR could not distinguish between guide
promotes the processing and loading of siRNAs/miRNAs onto
strands bound to Ago2 as a single strand or as part of an siRNA
RISCs by facilitating the interaction between TRBP and RNAs in
lishing Gr
duplex, we performed northern blot analysis using the same immu-
mammalian cells.
noprecipitated RNAs to determine the amounts of both guide-strand
To further confirm the role of TRBP in enoxacin-mediated RNAi-
and passenger-strand siRNAs associated with Ago2 (Fig. 4d, middle
enhancing activity, we performed a series of RNAi experiments using
panel). Upon addition of enoxacin, both guide and passenger strands
different amounts of siRNA duplexes against TRBP. The siRNAs
associated with Ago2 increased. Interestingly, the relative ratio
against TRBP were transfected into cells stably expressing an shRNA
between guide and passenger strands associated with Ago2 also
against GAPDH (Fig. 1c). We determined the RNAi-enhancing
increased by 30% in the presence of enoxacin, which suggests that
activity of enoxacin in the presence of different levels of TRBP
2008 Nature Pub
enoxacin could promote the loading of siRNA duplexes onto RISCs.
mRNA using the knockdown level of GAPDH as readout. Merely
reducing TRBP mRNA to 85% had no effect on shGAPDH-mediated
RNAi-enhancing activity is TRBP dependent
mRNA degradation (Fig. 5c and Supplementary Table 4); further-
Dicer and TRBP play critical roles in the processing and loading of
more, the reduction of TRBP mRNA to 50–80% had no effect on
miRNAs and siRNAs onto the RI. We next examined whether
RNAi-enhancing activity (Fig. 5c and Supplementary Table 4).
enoxacin might be involved in the processing mediated by Dicer and
However, when the TRBP mRNA level dropped below 22%, RNAi-
TRBP using an established in vitro processing assay. Enoxacin had no
enhancing activity was greatly reduced (Fig. 5c and Supplementary
effect on the processing of pre-let-7 or pre-miR-30a by Dicer alone.
Table 4). These results together suggest that the RNAi-enhancing
However, the addition of enoxacin could enhance the processing of
activity of enoxacin is TRBP dependent.
let-7 or pre-miR-30a by Dicer and TRBP together (Fig. 5a andSupplementary Fig. 6). This enhancement was not observed with
Enoxacin enhances RNAi in vivo
the addition of oxolinic acid, which has much less RNAi-enhancing
To determine whether enoxacin has similar effects in vivo, we tested it
activity (Fig. 3 and Supplementary Fig. 7a). This result indicated that
in a GFP transgenic mouse line. When a lentivirus expressing shGFP
enoxacin might target TRBP. Because TRBP binds to miRNA pre-
(Lv-siGFP) is injected into these mice early in development, the
cursors and facilitates the processing and loading of miRNAs, we
construct knocks-down GFP expressionHowever, injections of
performed a series of RNA-binding assays to examine the effect of
Lv-siGFP into adult mice did not alter GFP protein levels substantially
enoxacin on the interaction between TRBP and miRNA precursor. We
(data not shown), possibly due to the stability of the GFP protein.
found that the presence of enoxacin increased the binding affinity
We therefore chose to monitor the effects of enoxacin through
of TRBP for miRNA precursors; the Kd under normal conditions is
measurements of GFP mRNA after targeted injections of Lv-siGFP
221 nM, whereas the Kd in the presence of enoxacin is 94 nM (Fig. 5b
in young pups. We chose mouse ears for the injections, because they
and Supplementary Fig. 7b). These results suggest that enoxacin
are easily accessible; we could deliver the virus to a complete ear and
NATURE BIOTECHNOLOGY
ADVANCE ONLINE PUBLICATION
a much higher concentration (250 mM) than we used in this study
(50 mM), we examined genome-wide expressioWe observed
very few genes with altered expression and found no genes thatdisplayed consistent and substantial changes in the two cell lines
tested in our assay. We did, however, find that the processing of those
miRNAs whose precursors are abundant at steady state in cells was
promoted by enoxacin. Previous studies have shown that overexpres-
sion of miRNAs could produce substantial changes in the mRNA
Relative GFP mRNA level (%)
level. However, in those studies the expression levels of specific
miRNAs were elevated substantially, whereas in our assay we saw only
a mild approximately twofold increase of mature miRNAs in the cells
treated with enoxacin. Furthermore, how this mild increase of specific
Lv-siGFP + enoxacin
mature miRNAs affects the existing translational suppression remains
to be studied in more detail. We have found that the twofold increase
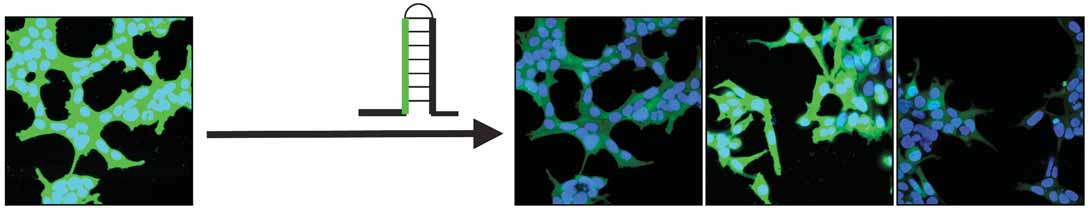
Figure 6 Enoxacin enhances RNAi in vivo. The ears of GFP transgenic mice
of specific mature miRNA had no effect on the translational suppres-
(ACTB-EGFP) were injected with shRNA-EGFP expressing lentivirus or
sion of corresponding reporter constructs (Supplementary Fig. 8).
Lv-siGFP, with or without enoxacin treatment (100 mM). The relative GFPmRNA levels in the control ears injected with Lv-siGFP only, injected with
Overall, these results indicate that the RNAi-enhancing effect of
both Lv-siGFP and enoxacin and injected with enoxacin alone are shown.
enoxacin is specific.
*P o 0.001 when Lv-siGFP is compared with control and Lv-siGFP+enoxacin
Using a series of in vitro and in vivo analyses, we found that the
is compared with Lv-siGFP alone.
enoxacin-mediated RNAi-enhancing activity is TRBP dependent, andenoxacin could facilitate the interaction between TRBP and RNAs.
obtain enough RNA from a single ear for quantitative RT-PCR
Furthermore, we found that enoxacin has no effect in an in vitro
analysis. In addition, we could compare the enoxacin-treated and
RISC-cleavage assay (Supplementary Fig. 9), which argues against the
enoxacin-untreated ears from the same mouse to reduce experimental
potential involvement of enoxacin in the step of mRNA-target
variation. Three groups of injections were performed: Lv-siGFP alone,
recognition and cleavage. Rather, these results together suggest that
Lv-siGFP with enoxacin and enoxacin alone. We performed multiple
enoxacin targets the step of RISC loading by enhancing the interaction
rounds of injections in 10-day-old mice and found that Lv-siGFP
between TRBP and RNAs. Indeed, it has been shown that the
alone reduced the GFP mRNA level to 80% of control tissues (20%
functionality of siRNAs is highly associated with the binding affinity
knockdown). The addition of enoxacin enhanced the knockdown
of TR; therefore the enhanced interaction between TRBP and
efficiency to 60% (40% GFP mRNA level remained), whereas enoxacin
RNAs mediated by enoxacin could be the basis of the RNAi-enhancing
alone had no effect on GFP expression (Fig. 6), which is consistent
activity. Our results indicate that TRBP plays an important role(s) in
with our previous cell culture data (Fig. 1b). These results suggest that
modulating the activity or efficacy of siRNAs, and enoxacin potentially
enoxacin enhances siRNA-mediated mRNA degradation in vivo.
increases RISC loading efficiency and enhances RNAi by targeting
lishing Gr
TRBP-RNA interactions. In summary, we have developed a novel cell-
based assay to monitor the activity of the RNAi pathway and identified
Small noncoding RNAs play important roles in animal development,
a small molecule that enhances siRNA-mediated mRNA degradation
evolution and human diseaAlthough our understanding of the
and promotes the biogenesis of endogenous miRNAs. Our results
mechanism of RNAi has dramatically increased with the identification
suggest that chemical screens could provide a powerful route to
of key proteins involved in the RNAi pathway, the modulation of this
understanding the modulation of the RNAi pathway. In addition, an
pathway in normal and disease states remains poorly understood,
RNAi enhancer could potentially facilitate the development of new
2008 Nature Pub
largely due to the overlapping, redundant and compensatory features
RNAi tools and therapeutics.
of the biological pathways regulated by small RNAs
Chemical biology approaches that use small molecules to perturb
protein networks of biological systems could be used to understand
DNA constructs, siRNAs, cell lines and transfections. Lv-siGFP was described
how the RNAi pathway is modulated. Using such an approach here,
previousThe short hairpin vectors shLuciferase and shGAPDH were
we identified a small molecule that enhances RNAi and promotes the
obtained from Open Biosystems. Details on other constructs and siRNA
biogenesis of miRNA by facilitating the interaction between TRBP and
duplexes are described in Supplementary Methods. All cell lines, includingHEK293, HeLa and NIH3T3, were cultured in DMEM with 10% FBS and
RNAs. Our results provide a proof-of-principle demonstration that
penicillin/streptomycin. Plasmids, 2-O-methyl-oligo siRNA and miRNA duplex
small molecules can be used to study the RNAi pathway.
were transfected into cells by Lipofectamine 2000 (Invitrogen). For BACE1
Enoxacin belongs to a family of synthetic antibacterial compounds,
siRNAs, the cells were trypsinized and reverse transfected with serial concen-
the fluoroquinoloneFluoroquinolones function as bacterial type II
trations of BACE1 siRNA (20 nM, 10 nM, 1 nM, 100 pM, 10 pM and 1 pM)
topoisomerase inhibitors. To determine the specificity of enoxacin as
using 0.2% Lipofectamine 2000 and the standard protocol. For siTRBP
an enhancer of RNAi, we also examined additional members of the
experiments, different concentrations of siTRBP (10 nM, 20 nM, 50 nM,
fluoroquinolone family; only a select few of these compounds
100 nM and 200 nM) were used. Stable cell lines were generated through
had substantial RNAi-enhancing activity, suggesting that the RNAi-
selection using appropriate antibiotics. All the transfection experiments were
enhancing activity does not depend on general fluoroquinolone
repeated at least three times. For most of the experiments performed in this
activity, but rather on the unique chemical structure of enoxacin
study, the final concentration of enoxacin is 50 mM. Enoxacin was incubatedwith cells for 48 h before biological assays.
and a few related family members.
To further exclude the possibility that the RNAi-enhancing effect we
Small-molecule screen, microscopy and fluorescent plate reading. Based on
observed is due to pleiotropic effects of enoxacin as a member of the
the levels of GFP fluorescence intensity, individual clones of 293-EGFP-siGFP
fluoroquinolone family, which is known to alter gene expression at
were isolated. Several clones were used for the chemical screen. Cells were
ADVANCE ONLINE PUBLICATION
NATURE BIOTECHNOLOGY
plated into 24-well plates. We added 2,000 individual drugs from The Spectrum
lysate proteins were separated by SDS-PAGE and probed with anti-Myc
Collection (10 mM in DMSO, MicroSource Discovery Systems) into individual
mAb to detect levels of Myc-tagged Ago2.
wells at a final concentration of 50 mM in 24 h. GFP fluorescence of each well
Purification of recombinant TRBP. His
was then visually inspected in 48 h using an inverted fluorescence microscope.
6-tagged TRBP was expressed in
Escherichia coli BL21 (DE3) cells and purified with the Ni-NTA Fast Start Kit
Compounds that obviously changed EGFP fluorescence were chosen for follow-
(Qiagen). The purity of the protein was analyzed by SDS-PAGE and its size was
up study. Images were taken using LSM 510 confocal microscope (Zeiss). To
compared with a His-tagged standard (Invitrogen). Protein concentration was
quantify RNAi-enhancing activity, we measured EGFP fluorescence intensity in
determined by Bradford Reagent Assay (Invitrogen).
48 h on an Analyst HT plate reader (Molecular Devices). An excitation filter at485 nm and an emission filter at 520 nm were used with a dichroic mirror of
In vitro miRNA precursor processing assay and TRBP binding assay. Let-7
505 nm. To calculate an EC50, the maximum effect of RNAi enhancement was
precursor RNA oligos were described previously and synthesized by Dhar-
defined as the enoxacin effect at 100 mM.
macon (ref. 30). Human miR-30a precursor (GCGACUGUAAACAUCCUCGACUGGAAGCUGUGAAGCCACAGAUGGGCUUUCAGUCGGAUGUUU
Chemical synthesis. Enoxacin and other fluoroquinolones tested here were
GCAGCUGC) was synthesized by Dharmacon, as well. They were radioactively
purchased from Sigma. Enoxacin-V1, V2 and V3 were chemically synthesized
labeled at the 5¢ end with 32P-g-ATP using T4 polynucleotide kinase (New
as detailed in Supplementary Methods.
England Biolabs). The labeled precursor was allowed to refold by heating at95 1C for 2 min, followed by incubation at 37 1C for 1 h. For the processing
Quantitative RT-PCRs, microarrays and western blot analyses. Real-time
assay, the labeled precursor was incubated with either Dicer or Dicer with
PCR was performed with gene-specific primers and Power SYBR Green PCR
TRBP in the buffer as described previously for 1 The reaction was
Master Mix (Applied Biosystems) using a 7500 Fast Real-Time PCR system
terminated and subjected to phenol/chloroform extraction and ethanol pre-
(Applied Biosystems). Profiling of mature miRNA expression was performed
cipitation. Then the samples were separated on a 15% TBE urea gel, transferred
using Applied Biosystems' TaqMan miRNA assays with 8-plex reverse tran-
and UV crosslinked to nylon membrane, which was then exposed for
scription and individual TaqMan miRNA real-time PCR assays according to
PhosphorImager scanning. Both unprocessed and processed precursors were
protocols provided by the vendor. Gene expression profiles were evaluated
quantified using Kodak MI software. The percentage of processed RNAs was
using Affymetrix Expression Arrays, HG-U133_Plus_2 and Mouse430_2. For
used to calculate relative cleavage activity, with no enoxacin as a control
western blotting, protein samples were separated on SDS-PAGE gels and then
(100%). Four independent experiments were performed and used to calculate
transferred to PVDF membranes (Millipore). Membranes were processed
mean and s.d.
following the ECL western blotting protocol (Amersham). The details of these
TRBP and let-7 precursor binding reactions were carried out in 1� binding
experiments are described in Supplementary Methods.
buffer (20 mM Tris-HCl, pH 8.0, 15 mM NaCl, 2.5 mM MgCl2) with 150 ng ofpurified recombinant TRBP and 10 ng 32P-labeled let-7 precursor in a total
Northern blot of small RNAs. RNAs were isolated with TRIzol (Invitrogen),
volume of 30 ml with or without 30 mM RNAi-E (or RNAi-E V5) at 30 1C for
and then separated on 15% TBE urea gel, transferred and UV crosslinked
5, 10, 30, 60, 90, 120 and 180 min. The reaction mixtures were then brought
to nylon membrane (Osmonics). 32P-UTP–labeled probes were prepared
into contact with a UV lamp in a CL-1000 Ultraviolet Crosslinker (Stratagene)
with the Ambion mirVana miRNA Probe Construction Kit. Membranes
for 5 min on ice. After crosslinking, the samples were mixed with an equal
were prehybridized at 65 1C for 1 h and hybridized for 12–16 h at 25 1C.
volume of 2� Gel Loading Buffer (Applied Biosystems) and incubated for
Membranes were then washed three times at 25 1C and two times at 42 1C.
5 min at 95 1C. The denatured samples were separated onto 10% SDS-PAGE
Membranes were exposed and scanned with a Typhoon 9200 PhosphorImager
followed by gel dry using Gel Dryer (Fisher). The dried gels were exposed to
lishing Gr
Storage Phosphor Screen (Amersham Pharmacia Biotech) and the screenswere scanned using a Typhoon 9200 PhosphorImager then quantified using
Determination of small RNA duplex loaded onto RISCs. To quantify the
Kodak MI software. The percentage of RNAs bound by TRBP was used to
transfected small RNA duplex loaded onto RISCs, we used the expression
calculate the binding activity. At least four independent experiments were
vector of Myc-Ago2 fusion protein and synthesized small RNA duplex. For ease
performed and used to calculate mean and s.d. For nitrocellulose filter binding
of quantification, we used duplexes that resemble human miR-125a. In an
assay, TRBP and let-7 precursor binding reactions were carried out in
earlier study we found that the expression of miR-125a in HEK293 cells is
1� binding buffer (20 mM Tris-HCl, pH 8.0, 15 mM NaCl, 2.5 mM MgCl2)
extremely low and could not be detected using either the MiRNA TaqMan assay
2008 Nature Pub
with different amounts of purified recombinant TRBP and 10 ng 32P-labeled
or northern blot. Briefly, HEK293FT (‘fast transfect') cells were either
let-7 precursor in a total volume of 30 ml with or without 30 mM enoxacin at
transfected with Myc-tagged Ago2 plasmid or cotransfected with both Myc-
30 1C for 60 min. Binding solutions were passed through MF-membrane filters
tagged Ago2 expression vector and miR-125a duplexes (100 mM) using
(0.45 HA, Millipore) and washed with 4 ml of ice-cold wash buffer containing
Lipofectamine 2000. The transfected cells were split 24 h after transfection
50 mM Tris and 20 mM KCl (pH7.4). After the wash buffer was drained,
and treated with either no enoxacin or 75-mM enoxacin for 48 h before
the membranes were dried for 2 min at 25 1C. The dried membranes were
collecting the cells for immunoprecipitation. The collected cells were lysed in
immersed into ScintiVerse BD Cocktail (FLUKA), and liquid scintillation
lysis buffer containing 20 mM HEPES, pH 7.4, 10 mM NaCl, 1 mM MgCl2,
was counted using a LS6500 Multipurpose Scintillation Counter (Beckman).
0.2 mM EDTA, 0.35% Triton-X100 (ref. 29) and 2� protease inhibitor cocktail
Data were plotted as relative amount of total RNA bound versus log of the
tablet (Roche) for 10 min. Ten percent of lysates were saved in 1 ml TRIzol for
TRBP concentration, and Kd was determined with KaleidaGraph software
total RNA isolation as inputs. The remaining lysates were centrifuged at 20,000g
(Synergy Software).
for 20 min at 4 1C. Protein concentrations of the supernatants were quantifiedusing Bradford Reagent (Bio-Rad). Lysates were adjusted to the concentrations
Mice and lentiviral injection. All animal procedures were performed based on
of 1 mg/ml, and 500 mg total proteins were used for immunoprecipitation
protocols approved by Emory University Institutional Animal Care and Use
overnight at 4 1C using anti-Myc mAb (Invitrogen) together with Protein A
Committee. The GFP transgenic line C57BL/6-Tg(ACTB-EGFP)1Osb/J was
Agarose Beads (Invitrogen). Immunoprecipitation beads were washed with the
obtained from The Jackson Laboratory. The lentivirus Lv-siGFP was produced
lysis buffer five times; 10% of the washed immunoprecipitation beads were
as described previouslyIndividual GFP transgenic mice (10 d old) were
used for western blots, and the remaining 90% for RNA extractions.
injected with the lentivirus Lv-siGFP (2 ml) into either one ear (to demonstrate
the effect of Lv-siGFP only) or both ears (for injection of enoxacin or control
with the High-Capacity cDNA Archive Kit (Applied Biosystems) combined
later) (day 10). Enoxacin or control solution (mock) (2 ml) was then injected
with hsa-miR-125a– and has-mir30a-3p–specific primers (Applied Biosystems).
once a day for 3 consecutive days (days 12, 13 and 14) into one of the ears 2 d
after Lv-siGFP injection. The concentration of injected enoxacin solution was
specific for has-miR-30a-3p and has-miR-125a. Proteins extracted from
100 mM. As a negative control, a group of mice were also injected with enoxacin
the 10% immunoprecipitation beads and an equal amount of input
or control solution into one ear. Mice were then killed (day 15), and the
NATURE BIOTECHNOLOGY
ADVANCE ONLINE PUBLICATION
ears were removed and used for RNA isolation and quantitative RT-PCR
6. Dykxhoorn, D.M. & Lieberman, J. Running interference: prospects and obstacles to
analysis of GFP mRNA. For each condition, at least six mice in three different
using small interfering RNAs as small molecule drugs. Annu. Rev. Biomed. Eng. 8,377–402 (2006).
groups were injected.
7. Tiscornia, G., Singer, O., Ikawa, M. & Verma, I.M. A general method for gene knockdown
in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl. Acad.
Statistical methods. We used single-factor ANOVA analysis to show significant
Sci. USA 100, 1844–1848 (2003).
differences between control and enoxacin treatment. We performed post-hoc
8. Hutvagner, G., Simard, M.J., Mello, C.C. & Zamore, P.D. Sequence-specific inhibition of
t-tests (two-sample assuming equal variances) to determine significance and
small RNA function. PLoS Biol. 2, E98 (2004).
indicated P value.
9. Patel, S.S. & Spencer, C.M. Enoxacin: a reappraisal of its clinical efficacy in the
treatment of genitourinary tract infections. Drugs 51, 137–160 (1996).
10. Pasquinelli, A.E. Demystifying small RNA pathways. Dev. Cell 10, 419–424 (2006).
Note: Supplementary information is available on the website.
11. Bhanot, S.K., Singh, M. & Chatterjee, N.R. The chemical and biological aspects of
fluoroquinolones: reality and dreams. Curr. Pharm. Des. 7, 311–335 (2001).
12. Hopper, D.C. & Wolfson, J.S. Quinolone Antimicrobial Agents 3rd edn. (ASM Press,
We would like to thank S. Warren, S. Chang, K. Garber and C. Strauss for
Washington, DC; 2003).
their helpful discussions and critical reading of the manuscript and H. Ju for
13. Mitscher, L.A. Bacterial topoisomerase inhibitors: quinolone and pyridone antibacterial
technical assistance. We thank I. Verma, J. Belasco and G. Hannon for providing
agents. Chem. Rev. 105, 559–592 (2005).
us the plasmids. Q.L. is a Damon Runyon Scholar (DRS-43) and is supported
14. Schaeffer, A.J. The expanding role of fluoroquinolones. Am. J. Med. 113 Suppl 1A,
45S–54S (2002).
by the Welch Foundation (I-1608). P.J. is supported by NIH grants (NS051630
15. Dalhoff, A. & Shalit, I. Immunomodulatory effects of quinolones. Lancet Infect. Dis. 3,
and MH076090). P.J. is the recipient of a Beckman Young Investigator Award
359–371 (2003).
and a Basil O'Connor Scholar Research Award and is an Alfred P. Sloan
16. Eriksson, E., Forsgren, A. & Riesbeck, K. Several gene programs are induced in
Research Fellow in Neuroscience.
ciprofloxacin-treated human lymphocytes as revealed by microarray analysis. J. Leukoc.
Biol. 74, 456–463 (2003).
AUTHOR CONTRIBUTIONS
17. Rand, T.A., Ginalski, K., Grishin, N.V. & Wang, X. Biochemical identification of
P.J. designed the research. G.S. conducted the chemical screen and identified the
Argonaute 2 as the sole protein required for RNA-induced silencing complex activity.
Proc. Natl. Acad. Sci. USA 101, 14385–14389 (2004).
small molecule presented in this paper. G.S. and Y.L. performed the majority of
18. Liu, J. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305,
mechanistic experiments. J.Z., L.L., Z.S. and C.H. performed chemical synthesis.
1437–1441 (2004).
G.S., Y.L., W.L., M.A.F., A.M.K. and C.W. performed additional testing on the
19. Bernstein, E., Caudy, A.A., Hammond, S.M. & Hannon, G.J. Role for a bidentate
compound. K.E.S. and R.D. performed miRNA profiling and developed the
ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001).
reporter system. A.W.S.C., Z.P. and Q.L. provided some reagents used in this
20. Chendrimada, T.P. et al. TRBP recruits the Dicer complex to Ago2 for microRNA
paper. G.S., Y.L. and P.J. wrote the paper.
processing and gene silencing. Nature 436, 740–744 (2005).
21. Jiang, F. et al. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes
COMPETING INTERESTS STATEMENT
Dev. 19, 1674–1679 (2005).
22. Forstemann, K. et al. Normal microRNA maturation and germ-line stem cell main-
The authors declare competing financial interests: details accompany the full-text
tenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS
HTML version of the paper at
Biol. 3, e236 (2005).
23. Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T. & Nishimune, Y. ‘Green mice' as a
Published online at
source of ubiquitous green cells. FEBS Lett. 407, 313–319 (1997).
Reprints and permissions information is available online at
24. Jackson, A.L. et al. Widespread siRNA ‘‘off-target'' transcript silencing mediated by
seed region sequence complementarity. RNA 12, 1179–1187 (2006).
25. Lim, L.P. et al. Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature 433, 769–773 (2005).
26. Jing, Q. et al. Involvement of microRNA in AU-rich element-mediated mRNA instabil-
1. Hannon, G.J. RNA interference. Nature 418, 244–251 (2002).
ity. Cell 120, 623–634 (2005).
lishing Gr
2. Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116,
27. Katoh, T. & Suzuki, T. Specific residues at every third position of siRNA shape its
281–297 (2004).
efficient RNAi activity. Nucleic Acids Res. 35, e27 (2007).
3. Plasterk, R.H. Micro RNAs in animal development. Cell 124, 877–881 (2006).
28. Duan, R., Pak, C. & Jin, P. Single nucleotide polymorphism associated with mature miR-
4. Zamore, P.D. & Haley, B. Ribo-gnome: the big world of small RNAs. Science 309,
125a alters the processing of pri-miRNA. Hum. Mol. Genet. 16, 1124–1131 (2007).
1519–1524 (2005).
29. Chu, C.Y. & Rana, T.M. Translation repression in human cells by microRNA-induced
5. Dykxhoorn, D.M. & Lieberman, J. The silent revolution: RNA interference as
gene silencing requires RCK/p54. PLoS Biol. 4, e210 (2006).
basic biology, research tool, and therapeutic. Annu. Rev. Med. 56, 401–423
30. Hutvagner, G. & Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme
complex. Science 297, 2056–2060 (2002).
2008 Nature Pub
ADVANCE ONLINE PUBLICATION
NATURE BIOTECHNOLOGY
Source: http://www.effigene.com/news/pdf/naturbiotech_2008.pdf
Nr. 45 Februar 2011 Magazin für Lehramtsanwärter/-innen Aus dem Inhalt: 3 Mathe unterrichten und den Lehrplanforderungen gerecht werden? 8 Kopiervorlagen aus PIK AS 11 Kreis und Winkel – eine Unterrichts- einheit für die Jahrgangsstufe 6 14 Junglehrer fordern Standards
An Information Brochure by The Gut Foundation Diverticular diseaseWhat is it?Diverticular disease affects the large bowel. The disease is usually confined to the sigmoid colon although it can involve all the colon. Diverticula are small pockets or sacs that protrude beyond the wall of the bowel and vary in size from that of a pinhead to a small grape. The mouth of the diverticulum is often narrow giving it a teardrop shape. The local bowel wall is thickened.











